A Toolbox for Herpesvirus miRNA Research: Construction of a Complete Set of KSHV miRNA Deletion Mutants
- PMID: 26907327
- PMCID: PMC4776209
- DOI: 10.3390/v8020054
A Toolbox for Herpesvirus miRNA Research: Construction of a Complete Set of KSHV miRNA Deletion Mutants
Abstract
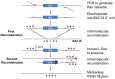
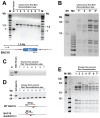
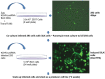
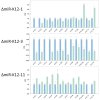
Kaposi's sarcoma-associated herpesvirus (KSHV) encodes 12 viral microRNAs (miRNAs) that are expressed during latency. Research into KSHV miRNA function has suffered from a lack of genetic systems to study viral miRNA mutations in the context of the viral genome. We used the Escherichia coli Red recombination system together with a new bacmid background, BAC16, to create mutants for all known KSHV miRNAs. The specific miRNA deletions or mutations and the integrity of the bacmids have been strictly quality controlled using PCR, restriction digestion, and sequencing. In addition, stable viral producer cell lines based on iSLK cells have been created for wildtype KSHV, for 12 individual miRNA knock-out mutants (ΔmiR-K12-1 through -12), and for mutants deleted for 10 of 12 (ΔmiR-cluster) or all 12 miRNAs (ΔmiR-all). NGS, in combination with SureSelect technology, was employed to sequence the entire latent genome within all producer cell lines. qPCR assays were used to verify the expression of the remaining viral miRNAs in a subset of mutants. Induction of the lytic cycle leads to efficient production of progeny viruses that have been used to infect endothelial cells. Wt BAC16 and miR mutant iSLK producer cell lines are now available to the research community.
Keywords: KSHV; bacmid; human herpesvirus 8; miRNA.
Figures

Similar articles
-
KSHV microRNAs: Tricks of the Devil.Trends Microbiol. 2017 Aug;25(8):648-661. doi: 10.1016/j.tim.2017.02.002. Epub 2017 Mar 2. Trends Microbiol. 2017. PMID: 28259385 Free PMC article. Review.
-
Modified Cross-Linking, Ligation, and Sequencing of Hybrids (qCLASH) Identifies Kaposi's Sarcoma-Associated Herpesvirus MicroRNA Targets in Endothelial Cells.J Virol. 2018 Mar 28;92(8):e02138-17. doi: 10.1128/JVI.02138-17. Print 2018 Apr 15. J Virol. 2018. PMID: 29386283 Free PMC article.
-
Epigenetic regulation of Kaposi's sarcoma-associated herpesvirus latency by virus-encoded microRNAs that target Rta and the cellular Rbl2-DNMT pathway.J Virol. 2010 Mar;84(6):2697-706. doi: 10.1128/JVI.01997-09. Epub 2010 Jan 13. J Virol. 2010. PMID: 20071580 Free PMC article.
-
Kaposi's sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells.Proc Natl Acad Sci U S A. 2005 Apr 12;102(15):5570-5. doi: 10.1073/pnas.0408192102. Epub 2005 Mar 30. Proc Natl Acad Sci U S A. 2005. PMID: 15800047 Free PMC article.
-
miRNAs and their roles in KSHV pathogenesis.Virus Res. 2019 Jun;266:15-24. doi: 10.1016/j.virusres.2019.03.024. Epub 2019 Apr 2. Virus Res. 2019. PMID: 30951791 Review.
Cited by
-
KSHV microRNAs: Tricks of the Devil.Trends Microbiol. 2017 Aug;25(8):648-661. doi: 10.1016/j.tim.2017.02.002. Epub 2017 Mar 2. Trends Microbiol. 2017. PMID: 28259385 Free PMC article. Review.
-
Herpesviruses shape tumour microenvironment through exosomal transfer of viral microRNAs.PLoS Pathog. 2017 Aug 24;13(8):e1006524. doi: 10.1371/journal.ppat.1006524. eCollection 2017 Aug. PLoS Pathog. 2017. PMID: 28837697 Free PMC article.
-
KSHV requires vCyclin to overcome replicative senescence in primary human lymphatic endothelial cells.PLoS Pathog. 2020 Jun 18;16(6):e1008634. doi: 10.1371/journal.ppat.1008634. eCollection 2020 Jun. PLoS Pathog. 2020. PMID: 32555637 Free PMC article.
-
Polypharmacology-based kinome screen identifies new regulators of KSHV reactivation.bioRxiv [Preprint]. 2023 Feb 1:2023.02.01.526589. doi: 10.1101/2023.02.01.526589. bioRxiv. 2023. Update in: PLoS Pathog. 2023 Sep 5;19(9):e1011169. doi: 10.1371/journal.ppat.1011169. PMID: 36778430 Free PMC article. Updated. Preprint.
-
Cis regulation within a cluster of viral microRNAs.Nucleic Acids Res. 2021 Sep 27;49(17):10018-10033. doi: 10.1093/nar/gkab731. Nucleic Acids Res. 2021. PMID: 34417603 Free PMC article.
References
-
- Soulier J., Grollet L., Oksenhendler E., Cacoub P., Cazals-Hatem D., Babinet P., d′Agay M.F., Clauvel J.P., Raphael M., Degos L., et al. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman′s disease. Blood. 1995;86:1276–1280. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

