Comprehensive Genomic Analysis and Expression Profiling of the NOX Gene Families under Abiotic Stresses and Hormones in Plants
- PMID: 26907500
- PMCID: PMC4824067
- DOI: 10.1093/gbe/evw035
Comprehensive Genomic Analysis and Expression Profiling of the NOX Gene Families under Abiotic Stresses and Hormones in Plants
Abstract
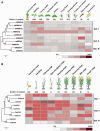
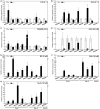
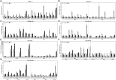
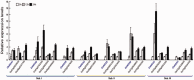
Plasma membrane NADPH oxidases (NOXs) are key producers of reactive oxygen species under both normal and stress conditions in plants and they form functional subfamilies. Studies of these subfamilies indicated that they show considerable evolutionary selection. We performed a comparative genomic analysis that identified 50 ferric reduction oxidases (FRO) and 77 NOX gene homologs from 20 species representing the eight major plant lineages within the supergroup Plantae: glaucophytes, rhodophytes, chlorophytes, bryophytes, lycophytes, gymnosperms, monocots, and eudicots. Phylogenetic and structural analysis classified these FRO and NOX genes into four well-conserved groups represented as NOX, FRO I, FRO II, and FRO III. Further analysis of NOXs of phylogenetic and exon/intron structures showed that single intron loss and gain had occurred, yielding the diversified gene structures during the evolution of NOXs family genes and which were classified into four conserved subfamilies which are represented as Sub.I, Sub.II, Sub.III, and Sub.IV. Additionally, both available global microarray data analysis and quantitative real-time PCR experiments revealed that the NOX genes in Arabidopsis and rice (Oryza sativa) have different expression patterns in different developmental stages, various abiotic stresses and hormone treatments. Finally, coexpression network analysis of NOX genes in Arabidopsis and rice revealed that NOXs have significantly correlated expression profiles with genes which are involved in plants metabolic and resistance progresses. All these results suggest that NOX family underscores the functional diversity and divergence in plants. This finding will facilitate further studies of the NOX family and provide valuable information for functional validation of this family in plants.
Keywords: NADPH oxidase (NOX); abiotic stress; coexpression; hormone; phylogenetic analysis.
© The Author 2016. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures




Similar articles
-
Genome-wide analysis of the NADK gene family in plants.PLoS One. 2014 Jun 26;9(6):e101051. doi: 10.1371/journal.pone.0101051. eCollection 2014. PLoS One. 2014. PMID: 24968225 Free PMC article.
-
Comparative in Silico Analysis of Ferric Reduction Oxidase (FRO) Genes Expression Patterns in Response to Abiotic Stresses, Metal and Hormone Applications.Molecules. 2018 May 12;23(5):1163. doi: 10.3390/molecules23051163. Molecules. 2018. PMID: 29757203 Free PMC article.
-
Regulation of ATG6/Beclin-1 homologs by abiotic stresses and hormones in rice (Oryza sativa L.).Genet Mol Res. 2012 Oct 11;11(4):3676-87. doi: 10.4238/2012.August.17.3. Genet Mol Res. 2012. PMID: 22930426
-
The Fundamental Role of NOX Family Proteins in Plant Immunity and Their Regulation.Int J Mol Sci. 2016 May 27;17(6):805. doi: 10.3390/ijms17060805. Int J Mol Sci. 2016. PMID: 27240354 Free PMC article. Review.
-
Structure, regulation and evolution of Nox-family NADPH oxidases that produce reactive oxygen species.FEBS J. 2008 Jul;275(13):3249-77. doi: 10.1111/j.1742-4658.2008.06488.x. Epub 2008 May 30. FEBS J. 2008. PMID: 18513324 Review.
Cited by
-
Improving crop performance under drought - cross-fertilization of disciplines.J Exp Bot. 2017 Mar 1;68(7):1393-1398. doi: 10.1093/jxb/erx042. J Exp Bot. 2017. PMID: 28338855 Free PMC article. No abstract available.
-
Genome-wide mining of respiratory burst homologs and its expression in response to biotic and abiotic stresses in Triticum aestivum.Genes Genomics. 2019 Sep;41(9):1027-1043. doi: 10.1007/s13258-019-00821-x. Epub 2019 May 28. Genes Genomics. 2019. PMID: 31140145
-
The Role of Endoplasmic Reticulum Stress Response in Pollen Development and Heat Stress Tolerance.Front Plant Sci. 2021 Apr 14;12:661062. doi: 10.3389/fpls.2021.661062. eCollection 2021. Front Plant Sci. 2021. PMID: 33936150 Free PMC article. Review.
-
Genome-wide identification and expression analysis of NADPH oxidase genes in response to ABA and abiotic stresses, and in fibre formation in Gossypium.PeerJ. 2020 Jan 17;8:e8404. doi: 10.7717/peerj.8404. eCollection 2020. PeerJ. 2020. PMID: 31988810 Free PMC article.
-
Structural complexity and functional diversity of plant NADPH oxidases.Amino Acids. 2018 Jan;50(1):79-94. doi: 10.1007/s00726-017-2491-5. Epub 2017 Oct 25. Amino Acids. 2018. PMID: 29071531 Free PMC article.
References
-
- Aguirre J, Rios-Momberg M, Hewitt D, Hansberg W. 2005. Reactive oxygen species and development in microbial eukaryotes. Trends Microbiol. 13:111–118. - PubMed
-
- Amicucci E, Gaschler K, Ward JM. 1999. NADPH oxidase genes from tomato (Lycopersicon esculentum) and curly-leaf pondweed (Potamogeton crispus). Plant Biol. 1:524–528.
-
- Anderson A, Bothwell JH, Laohavisit A, Smith AG, Davies JM. 2011. NOX or not? Evidence for algal NADPH oxidases. Trends Plant Sci. 16: 579–581. - PubMed
-
- Arthikala MK, et al. 2014. RbohB, a Phaseolus vulgaris NADPH oxidase gene, enhances symbiosome number, bacteroid size, and nitrogen fixation in nodules and impairs mycorrhizal colonization. New Phytol. 202:886–900. - PubMed
-
- Asano T, et al. 2012. A rice calcium-dependent protein kinase OsCPK12 oppositely modulates salt-stress tolerance and blast disease resistance. Plant J. 69:26–36. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

