SIRT7 Is Activated by DNA and Deacetylates Histone H3 in the Chromatin Context
- PMID: 26907567
- PMCID: PMC4850736
- DOI: 10.1021/acschembio.5b01084
SIRT7 Is Activated by DNA and Deacetylates Histone H3 in the Chromatin Context
Abstract
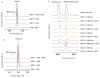
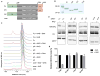
Mammalian sirtuins (SIRT1-7) are members of a highly conserved family of nicotinamide adenine dinucleotide (NAD(+))-dependent protein deacetylases that regulate many biological processes including metabolism, genome stability, and transcription. Among the seven human sirtuins, SIRT7 is the least understood, to a large extent due to the lack of enzymatic activity in vitro. Here, we reported that SIRT7 can be activated by DNA to hydrolyze the acetyl group from lysine residues in vitro on histone peptides and histones in the chromatin context. Both N- and C- termini of SIRT7 are important for the DNA-activated deacetylase activity. The regulatory mechanism of SIRT7 is different from that of SIRT6, which also showed increased activity on chromatin substrates, but the deacetylase activity of SIRT6 on a peptide substrate cannot be activated by DNA. This finding provides an improved enzymatic activity assay of SIRT7 that will promote the development of SIRT7 modulators. Further investigation into the activation mechanism of SIRT7 by DNA could provide new insights into its biological function and help the development of sirtuin activators.
Figures

Similar articles
-
Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by mammalian sirtuins.J Biol Chem. 2013 Oct 25;288(43):31350-6. doi: 10.1074/jbc.C113.511261. Epub 2013 Sep 18. J Biol Chem. 2013. PMID: 24052263 Free PMC article.
-
SIRT7 Is an RNA-Activated Protein Lysine Deacylase.ACS Chem Biol. 2017 Jan 20;12(1):300-310. doi: 10.1021/acschembio.6b00954. Epub 2016 Dec 20. ACS Chem Biol. 2017. PMID: 27997115 Free PMC article.
-
SIRT7 is a histone desuccinylase that functionally links to chromatin compaction and genome stability.Nat Commun. 2016 Jul 20;7:12235. doi: 10.1038/ncomms12235. Nat Commun. 2016. PMID: 27436229 Free PMC article.
-
Sirtuins and DNA damage repair: SIRT7 comes to play.Nucleus. 2017 Mar 4;8(2):107-115. doi: 10.1080/19491034.2016.1264552. Epub 2017 Feb 17. Nucleus. 2017. PMID: 28406750 Free PMC article. Review.
-
Mammalian Sirtuins SIRT4 and SIRT7.Prog Mol Biol Transl Sci. 2018;154:147-168. doi: 10.1016/bs.pmbts.2017.11.001. Epub 2018 Jan 19. Prog Mol Biol Transl Sci. 2018. PMID: 29413176 Review.
Cited by
-
Age-Associated Changes of Sirtuin 2 Expression in CNS and the Periphery.Biology (Basel). 2023 Nov 29;12(12):1476. doi: 10.3390/biology12121476. Biology (Basel). 2023. PMID: 38132302 Free PMC article. Review.
-
Updates on the epigenetic roles of sirtuins.Curr Opin Chem Biol. 2019 Aug;51:18-29. doi: 10.1016/j.cbpa.2019.01.023. Epub 2019 Mar 12. Curr Opin Chem Biol. 2019. PMID: 30875552 Free PMC article. Review.
-
Potent Activation of NAD+-Dependent Deacetylase Sirt7 by Nucleosome Binding.ACS Chem Biol. 2022 Aug 19;17(8):2248-2261. doi: 10.1021/acschembio.2c00348. Epub 2022 Aug 8. ACS Chem Biol. 2022. PMID: 35939806 Free PMC article.
-
Photo Cross-Linking Probes Containing ϵ-N-Thioacyllysine and ϵ-N-Acyl-(δ-aza)lysine Residues.Chemistry. 2020 Mar 23;26(17):3862-3869. doi: 10.1002/chem.201905338. Epub 2020 Mar 3. Chemistry. 2020. PMID: 31922630 Free PMC article.
-
SIRT7 Is a Lysine Deacylase with a Preference for Depropionylation and Demyristoylation.Int J Mol Sci. 2025 Mar 28;26(7):3153. doi: 10.3390/ijms26073153. Int J Mol Sci. 2025. PMID: 40243935 Free PMC article.
References
-
- Imai S-i, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. - PubMed
-
- Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun. 2000;273:793–798. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

