Hfq: the flexible RNA matchmaker
- PMID: 26907610
- PMCID: PMC4821791
- DOI: 10.1016/j.mib.2016.02.003
Hfq: the flexible RNA matchmaker
Abstract
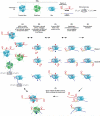
The RNA chaperone protein Hfq is critical to the function of small, base pairing RNAs in many bacteria. In the past few years, structures and modeling of wild type Hfq and assays of various mutants have documented that the homohexameric Hfq ring can contact RNA at four sites (proximal face, distal face, rim and C-terminal tail) and that different RNAs bind to these sites in various configurations. These studies together with novel in vitro and in vivo experimental approaches are beginning to give mechanistic insights into how Hfq acts to promote small RNA-mRNA pairing and indicate that flexibility is integral to the Hfq role in RNA matchmaking.
Published by Elsevier Ltd.
Figures

References
-
- Franze de Fernandez MT, Eoyang L, August JT. Factor fraction required for the synthesis of bacteriophage Qβ-RNA. Nature. 1968;219:588–590. - PubMed
-
- Sobrero P, Valverde C. The bacterial protein Hfq: much more than a mere RNA-binding factor. Crit. Rev. Microbiol. 2012;38:276–299. [This very comprehensive review summarizes information about the surface charge of Hfq from different bacteria, known hfq deletion phenotypes, and the controversy surrounding the role of the C-terminal tail.] - PubMed
-
- Valentin-Hansen P, Eriksen M, Udesen C. The bacterial Sm-like protein Hfq: a key player in RNA transactions. Mol. Microbiol. 2004;51:1525–1533. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

