Distal Alternative Last Exons Localize mRNAs to Neural Projections
- PMID: 26907613
- PMCID: PMC4798900
- DOI: 10.1016/j.molcel.2016.01.020
Distal Alternative Last Exons Localize mRNAs to Neural Projections
Abstract
Spatial restriction of mRNA to distinct subcellular locations enables local regulation and synthesis of proteins. However, the organizing principles of mRNA localization remain poorly understood. Here we analyzed subcellular transcriptomes of neural projections and soma of primary mouse cortical neurons and two neuronal cell lines and found that alternative last exons (ALEs) often confer isoform-specific localization. Surprisingly, gene-distal ALE isoforms were four times more often localized to neurites than gene-proximal isoforms. Localized isoforms were induced during neuronal differentiation and enriched for motifs associated with muscleblind-like (Mbnl) family RNA-binding proteins. Depletion of Mbnl1 and/or Mbnl2 reduced localization of hundreds of transcripts, implicating Mbnls in localization of mRNAs to neurites. We provide evidence supporting a model in which the linkage between genomic position of ALEs and subcellular localization enables coordinated induction of localization-competent mRNA isoforms through a post-transcriptional regulatory program that is induced during differentiation and reversed in cellular reprogramming and cancer.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures


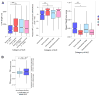
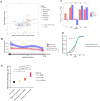

References
-
- Andreassi C, Riccio A. To localize or not to localize: mRNA fate is in 3′UTR ends. Trends Cell Biol. 2009;19:465–474. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

