Generation of an algorithm based on minimal gene sets to clinically subtype triple negative breast cancer patients
- PMID: 26908167
- PMCID: PMC4763445
- DOI: 10.1186/s12885-016-2198-0
Generation of an algorithm based on minimal gene sets to clinically subtype triple negative breast cancer patients
Erratum in
-
Erratum to: Generation of an algorithm based on minimal gene sets to clinically subtype triple negative breast cancer patients.BMC Cancer. 2016 Apr 18;16:275. doi: 10.1186/s12885-016-2307-0. BMC Cancer. 2016. PMID: 27090641 Free PMC article. No abstract available.
Abstract
Background: Recently, a gene expression algorithm, TNBCtype, was developed that can divide triple-negative breast cancer (TNBC) into molecularly-defined subtypes. The algorithm has potential to provide predictive value for TNBC subtype-specific response to various treatments. TNBCtype used in a retrospective analysis of neoadjuvant clinical trial data of TNBC patients demonstrated that TNBC subtype and pathological complete response to neoadjuvant chemotherapy were significantly associated. Herein we describe an expression algorithm reduced to 101 genes with the power to subtype TNBC tumors similar to the original 2188-gene expression algorithm and predict patient outcomes.
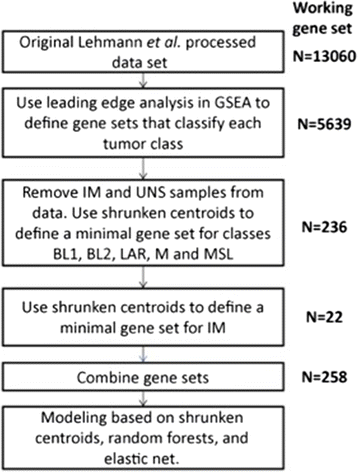
Methods: The new classification model was built using the same expression data sets used for the original TNBCtype algorithm. Gene set enrichment followed by shrunken centroid analysis were used for feature reduction, then elastic-net regularized linear modeling was used to identify genes for a centroid model classifying all subtypes, comprised of 101 genes. The predictive capability of both this new "lean" algorithm and the original 2188-gene model were applied to an independent clinical trial cohort of 139 TNBC patients treated initially with neoadjuvant doxorubicin/cyclophosphamide and then randomized to receive either paclitaxel or ixabepilone to determine association of pathologic complete response within the subtypes.
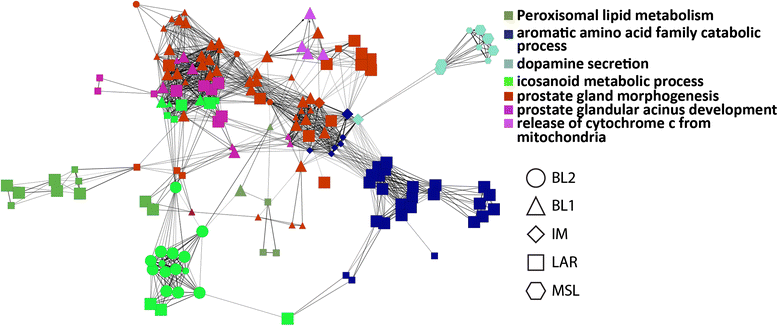
Results: The new 101-gene expression model reproduced the classification provided by the 2188-gene algorithm and was highly concordant in the same set of seven TNBC cohorts used to generate the TNBCtype algorithm (87%), as well as in the independent clinical trial cohort (88%), when cases with significant correlations to multiple subtypes were excluded. Clinical responses to both neoadjuvant treatment arms, found BL2 to be significantly associated with poor response (Odds Ratio (OR) =0.12, p=0.03 for the 2188-gene model; OR = 0.23, p < 0.03 for the 101-gene model). Additionally, while the BL1 subtype trended towards significance in the 2188-gene model (OR = 1.91, p = 0.14), the 101-gene model demonstrated significant association with improved response in patients with the BL1 subtype (OR = 3.59, p = 0.02).
Conclusions: These results demonstrate that a model using small gene sets can recapitulate the TNBC subtypes identified by the original 2188-gene model and in the case of standard chemotherapy, the ability to predict therapeutic response.
Figures
References
-
- Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, Lickley LA, Rawlinson E, Sun P, Narod SA. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res. 2007;13(15 Pt 1):4429–34. - PubMed
-
- Liedtke C, Mazouni C, Hess KR, André F, Tordai A, Mejia JA, Symmans WF, et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol. 2008;26(8):1275–81. - PubMed
-
- Wang-Lopeza Q, Chalabia N, Abriala C, Radosevic-Robina N, Durandoa X, Mouret-Reyniera MA, et al. Can pathologic complete response (pCR) be used as a surrogate marker of survival after neoadjuvant therapy for breast cancer? Crit Rev Oncol Hematol. 2015;95:88–104. doi: 10.1016/j.critrevonc.2015.02.011. - DOI - PubMed
-
- Guidance for industry: pathological complete response in neoadjuvant treatment of high-risk early-stage breast cancer: use as an endpoint to support accelerated approval. http://www.fda.gov/downloads/drugs/guidancecomplianceregulatoryinformati....
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

