An overview of macroautophagy in yeast
- PMID: 26908221
- PMCID: PMC4846508
- DOI: 10.1016/j.jmb.2016.02.021
An overview of macroautophagy in yeast
Abstract
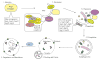
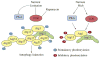
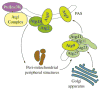
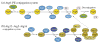
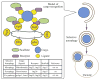
Macroautophagy is an evolutionarily conserved dynamic pathway that functions primarily in a degradative manner. A basal level of macroautophagy occurs constitutively, but this process can be further induced in response to various types of stress including starvation, hypoxia and hormonal stimuli. The general principle behind macroautophagy is that cytoplasmic contents can be sequestered within a transient double-membrane organelle, an autophagosome, which subsequently fuses with a lysosome or vacuole (in mammals, or yeast and plants, respectively), allowing for degradation of the cargo followed by recycling of the resulting macromolecules. Through this basic mechanism, macroautophagy has a critical role in cellular homeostasis; however, either insufficient or excessive macroautophagy can seriously compromise cell physiology, and thus, it needs to be properly regulated. In fact, a wide range of diseases are associated with dysregulation of macroautophagy. There has been substantial progress in understanding the regulation and molecular mechanisms of macroautophagy in different organisms; however, many questions concerning some of the most fundamental aspects of macroautophagy remain unresolved. In this review, we summarize current knowledge about macroautophagy mainly in yeast, including the mechanism of autophagosome biogenesis, the function of the core macroautophagic machinery, the regulation of macroautophagy and the process of cargo recognition in selective macroautophagy, with the goal of providing insights into some of the key unanswered questions in this field.
Keywords: autophagosome biogenesis; autophagy; cargo recognition; regulation; stress.
Copyright © 2016 Elsevier Ltd. All rights reserved.
Figures

References
-
- Klionsky DJ, Cregg JM, Dunn WA, Jr, Emr SD, Sakai Y, Sandoval IV, et al. A unified nomenclature for yeast autophagy-related genes. Dev Cell. 2003;5:539–45. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

