Host-Primed Ebola Virus GP Exposes a Hydrophobic NPC1 Receptor-Binding Pocket, Revealing a Target for Broadly Neutralizing Antibodies
- PMID: 26908579
- PMCID: PMC4791852
- DOI: 10.1128/mBio.02154-15
Host-Primed Ebola Virus GP Exposes a Hydrophobic NPC1 Receptor-Binding Pocket, Revealing a Target for Broadly Neutralizing Antibodies
Abstract
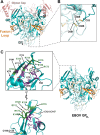
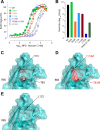
The filovirus surface glycoprotein (GP) mediates viral entry into host cells. Following viral internalization into endosomes, GP is cleaved by host cysteine proteases to expose a receptor-binding site (RBS) that is otherwise hidden from immune surveillance. Here, we present the crystal structure of proteolytically cleaved Ebola virus GP to a resolution of 3.3 Å. We use this structure in conjunction with functional analysis of a large panel of pseudotyped viruses bearing mutant GP proteins to map the Ebola virus GP endosomal RBS at molecular resolution. Our studies indicate that binding of GP to its endosomal receptor Niemann-Pick C1 occurs in two distinct stages: the initial electrostatic interactions are followed by specific interactions with a hydrophobic trough that is exposed on the endosomally cleaved GP1 subunit. Finally, we demonstrate that monoclonal antibodies targeting the filovirus RBS neutralize all known filovirus GPs, making this conserved pocket a promising target for the development of panfilovirus therapeutics.
Importance: Ebola virus uses its glycoprotein (GP) to enter new host cells. During entry, GP must be cleaved by human enzymes in order for receptor binding to occur. Here, we provide the crystal structure of the cleaved form of Ebola virus GP. We demonstrate that cleavage exposes a site at the top of GP and that this site binds the critical domain C of the receptor, termed Niemann-Pick C1 (NPC1). We perform mutagenesis to find parts of the site essential for binding NPC1 and map distinct roles for an upper, charged crest and lower, hydrophobic trough in cleaved GP. We find that this 3-dimensional site is conserved across the filovirus family and that antibody directed against this site is able to bind cleaved GP from every filovirus tested and neutralize viruses bearing those GPs.
Copyright © 2016 Bornholdt et al.
Figures

References
-
- Negredo A, Palacios G, Vázquez-Morón S, González F, Dopazo H, Molero F, Juste J, Quetglas J, Savji N, de la Cruz Martínez M, Herrera JE, Pizarro M, Hutchison SK, Echevarría JE, Lipkin WI, Tenorio A. 2011. Discovery of an ebolavirus-like filovirus in Europe. PLoS Pathog 7:e1002304. doi:10.1371/journal.ppat.1002304. - DOI - PMC - PubMed
-
- Baize S, Pannetier D, Oestereich L, Rieger T, Koivogui L, Magassouba N, Soropogui B, Sow MS, Keïta S, De Clerck H, Tiffany A, Dominguez G, Loua M, Traoré A, Kolié M, Malano ER, Heleze E, Bocquin A, Mély S, Raoul H, Caro V, Cadar D, Gabriel M, Pahlmann M, Tappe D, Schmidt-Chanasit J, Impouma B, Diallo AK, Formenty P, Van Herp M, Gunther S. 2014. Emergence of Zaire Ebola virus disease in Guinea. N Engl J Med 371:1418–1425. doi:10.1056/NEJMoa1404505. - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

