The Non-structural Protein 5 and Matrix Protein Are Antigenic Targets of T Cell Immunity to Genotype 1 Porcine Reproductive and Respiratory Syndrome Viruses
- PMID: 26909080
- PMCID: PMC4755262
- DOI: 10.3389/fimmu.2016.00040
The Non-structural Protein 5 and Matrix Protein Are Antigenic Targets of T Cell Immunity to Genotype 1 Porcine Reproductive and Respiratory Syndrome Viruses
Abstract
The porcine reproductive and respiratory syndrome virus (PRRSV) is the cause of one of the most economically important diseases affecting swine worldwide. Efforts to develop a next-generation vaccine have largely focused on envelope glycoproteins to target virus-neutralizing antibody responses. However, these approaches have failed to demonstrate the necessary efficacy to progress toward market. T cells are crucial to the control of many viruses through cytolysis and cytokine secretion. Since control of PRRSV infection is not dependent on the development of neutralizing antibodies, it has been proposed that T cell-mediated immunity plays a key role. Therefore, we hypothesized that conserved T cell antigens represent prime candidates for the development a novel PRRS vaccine. Antigens were identified by screening a proteome-wide synthetic peptide library with T cells from cohorts of pigs rendered immune by experimental infections with a closely related (subtype 1) or divergent (subtype 3) PRRSV-1 strain. Dominant T cell IFN-γ responses were directed against the non-structural protein 5 (NSP5), and to a lesser extent, the matrix (M) protein. The majority of NSP5-specific CD8 T cells and M-specific CD4 T cells expressed a putative effector memory phenotype and were polyfunctional as assessed by coexpression of TNF-α and mobilization of the cytotoxic degranulation marker CD107a. Both antigens were generally well conserved among strains of both PRRSV genotypes. Thus, M and NSP5 represent attractive vaccine candidate T cell antigens, which should be evaluated further in the context of PRRSV vaccine development.
Keywords: IFN-γ; T cell; antigen identification; phenotype and function; porcine reproductive and respiratory syndrome virus; vaccine.
Figures

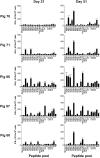
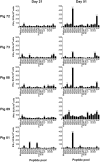
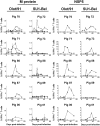
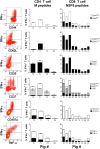

References
-
- Holtkamp DJ, Kliebenstein JB, Neumann EJ, Mowrer C, Haley C. Assessment of the economic impact of porcine reproductive and respiratory syndrome virus on United States pork producers. J Swine Health Prod (2013) 21:72–84.
-
- Morgan SB, Graham SP, Salguero FJ, Sanchez Cordon PJ, Mokhtar H, Rebel JM, et al. Increased pathogenicity of European porcine reproductive and respiratory syndrome virus is associated with enhanced adaptive responses and viral clearance. Vet Microbiol (2013) 163:13–22.10.1016/j.vetmic.2012.11.024 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

