Activation of the methylation cycle in cells reprogrammed into a stem cell-like state
- PMID: 26909364
- PMCID: PMC4735514
- DOI: 10.18632/oncoscience.280
Activation of the methylation cycle in cells reprogrammed into a stem cell-like state
Abstract
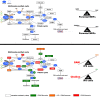
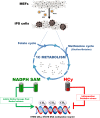
Generation of induced pluripotent stem (iPS) cells and cancer biogenesis share similar metabolic switches. Most studies have focused on how the establishment of a cancer-like glycolytic phenotype is necessary for the optimal routing of somatic cells for achieving stemness. However, relatively little effort has been dedicated towards elucidating how one-carbon (1C) metabolism is retuned during acquisition of stem cell identity. Here we used ultra-high pressure liquid chromatography coupled to an electrospray ionization source and a triple-quadrupole mass spectrometer [UHPLC-ESI-QqQ-MS/MS] to quantitatively examine the methionine/folate bi-cyclic 1C metabolome during nuclear reprogramming of somatic cells into iPS cells. iPS cells optimize the synthesis of the universal methyl donor S-adenosylmethionine (SAM), apparently augment the ability of the redox balance regulator NADPH in SAM biosynthesis, and greatly increase their methylation potential by triggering a high SAM:S-adenosylhomocysteine (SAH) ratio. Activation of the methylation cycle in iPS cells efficiently prevents the elevation of homocysteine (Hcy), which could alter global DNA methylation and induce mitochondrial toxicity, oxidative stress and inflammation. In this regard, the methyl donor choline is also strikingly accumulated in iPS cells, suggesting perhaps an overactive intersection of the de novo synthesis of choline with the methionine-Hcy cycle. Activation of methylogenesis and maintenance of an optimal SAM:Hcy ratio might represent an essential function of 1C metabolism to provide a labile pool of methyl groups and NADPH-dependent redox products required for successfully establishing and maintaining an embryonic-like DNA methylation imprint in stem cell states.
Keywords: Homocysteine; S-adenosylhomocysteine; iPS cells; one-carbon metabolism; stem cells.
Conflict of interest statement
There are no conflicts of interest to disclose.
Figures

References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

