Genome-wide functional genetic screen with the anticancer agent AMPI-109 identifies PRL-3 as an oncogenic driver in triple-negative breast cancers
- PMID: 26909599
- PMCID: PMC4941275
- DOI: 10.18632/oncotarget.7462
Genome-wide functional genetic screen with the anticancer agent AMPI-109 identifies PRL-3 as an oncogenic driver in triple-negative breast cancers
Abstract
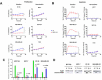
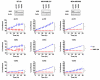
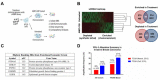
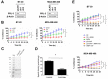
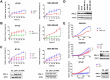
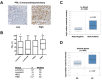
Triple-negative breast cancers (TNBC) are among the most aggressive and heterogeneous cancers with a high propensity to invade, metastasize and relapse. Here, we demonstrate that the anticancer compound, AMPI-109, is selectively efficacious in inhibiting proliferation and inducing apoptosis of multiple TNBC subtype cell lines as assessed by activation of pro-apoptotic caspases-3 and 7, PARP cleavage and nucleosomal DNA fragmentation. AMPI-109 had little to no effect on growth in the majority of non-TNBC cell lines examined. We therefore utilized AMPI-109 in a genome-wide shRNA screen in the TNBC cell line, BT-20, to investigate the utility of AMPI-109 as a tool in helping to identify molecular alterations unique to TNBC. Our screen identified the oncogenic phosphatase, PRL-3, as a potentially important driver of TNBC growth, migration and invasion. Through stable lentiviral knock downs and transfection with catalytically impaired PRL-3 in TNBC cells, loss of PRL-3 expression, or functionality, led to substantial growth inhibition. Moreover, AMPI-109 treatment, downregulation of PRL-3 expression or impairment of PRL-3 activity reduced TNBC cell migration and invasion. Histological evaluation of human breast cancers revealed PRL-3 was significantly, though not exclusively, associated with the TNBC subtype and correlated positively with regional and distant metastases, as well as 1 and 3 year relapse free survival. Collectively, our study is proof-of-concept that AMPI-109, a selectively active agent against TNBC cell lines, can be used as a molecular tool to uncover unique drivers of disease progression, such as PRL-3, which we show promotes oncogenic phenotypes in TNBC cells.
Keywords: AMPI-109; PRL-3; functional genomics; phosphatase; triple-negative breast cancer.
Conflict of interest statement
The authors disclose no potential conflicts of interest.
Figures
Similar articles
-
PRL-3 engages the focal adhesion pathway in triple-negative breast cancer cells to alter actin structure and substrate adhesion properties critical for cell migration and invasion.Cancer Lett. 2016 Oct 1;380(2):505-512. doi: 10.1016/j.canlet.2016.07.017. Epub 2016 Jul 21. Cancer Lett. 2016. PMID: 27452906 Free PMC article.
-
Phosphatase PTP4A3 Promotes Triple-Negative Breast Cancer Growth and Predicts Poor Patient Survival.Cancer Res. 2016 Apr 1;76(7):1942-53. doi: 10.1158/0008-5472.CAN-14-0673. Epub 2016 Feb 26. Cancer Res. 2016. PMID: 26921331 Free PMC article.
-
Identification of selective cytotoxic and synthetic lethal drug responses in triple negative breast cancer cells.Mol Cancer. 2016 May 10;15(1):34. doi: 10.1186/s12943-016-0517-3. Mol Cancer. 2016. PMID: 27165605 Free PMC article.
-
Sequential combination of docetaxel with a SHP-1 agonist enhanced suppression of p-STAT3 signaling and apoptosis in triple negative breast cancer cells.J Mol Med (Berl). 2017 Sep;95(9):965-975. doi: 10.1007/s00109-017-1549-x. Epub 2017 Jun 4. J Mol Med (Berl). 2017. PMID: 28578456
-
STAT3 as a potential therapeutic target in triple negative breast cancer: a systematic review.J Exp Clin Cancer Res. 2019 May 14;38(1):195. doi: 10.1186/s13046-019-1206-z. J Exp Clin Cancer Res. 2019. PMID: 31088482 Free PMC article.
Cited by
-
Expression of phosphatase of regenerating liver (PRL)-3, is independently associated with biochemical failure, clinical failure and death in prostate cancer.PLoS One. 2017 Nov 30;12(11):e0189000. doi: 10.1371/journal.pone.0189000. eCollection 2017. PLoS One. 2017. PMID: 29190795 Free PMC article.
-
PRL-3: unveiling a new horizon in cancer therapy.Acta Pharmacol Sin. 2025 May 8. doi: 10.1038/s41401-025-01563-1. Online ahead of print. Acta Pharmacol Sin. 2025. PMID: 40341216 Review.
-
Targeting ovarian cancer and endothelium with an allosteric PTP4A3 phosphatase inhibitor.Oncotarget. 2017 Dec 30;9(9):8223-8240. doi: 10.18632/oncotarget.23787. eCollection 2018 Feb 2. Oncotarget. 2017. PMID: 29492190 Free PMC article.
-
PRL3 as a therapeutic target for novel cancer immunotherapy in multiple cancer types.Theranostics. 2023 Mar 21;13(6):1876-1891. doi: 10.7150/thno.79265. eCollection 2023. Theranostics. 2023. PMID: 37064866 Free PMC article. Review.
-
Loss of the oncogenic phosphatase PRL-3 promotes a TNF-R1 feedback loop that mediates triple-negative breast cancer growth.Oncogenesis. 2016 Aug 15;5(8):e255. doi: 10.1038/oncsis.2016.50. Oncogenesis. 2016. PMID: 27526109 Free PMC article.
References
-
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl Acad. Sci. 2001;98:10869–10874. - PMC - PubMed
-
- Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, Lickley LA, Rawlinson E, Sun P, Narod SA. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res. 2007;13:4429–4434. - PubMed
-
- Hudis CA, Gianni L. Triple-negative breast cancer: an unmet medical need. Oncologist. 2011;16(Suppl 1):1–11. - PubMed
-
- Villarreal-Garza C, Khalaf D, Bouganim N, Clemons M, Pena-Curiel O, Baez-Revueltas, Kiss A, Kassan F, Enright K, Verma S, Pritchard K, Myers J, Dent R. Platinum-based chemotherapy in triple-negative advanced breast cancer. Breast Cancer Res. Treat. 2014;146:567–572. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

