DNAH11 Localization in the Proximal Region of Respiratory Cilia Defines Distinct Outer Dynein Arm Complexes
- PMID: 26909801
- PMCID: PMC4979367
- DOI: 10.1165/rcmb.2015-0353OC
DNAH11 Localization in the Proximal Region of Respiratory Cilia Defines Distinct Outer Dynein Arm Complexes
Abstract
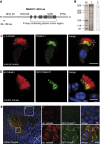
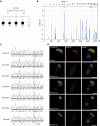
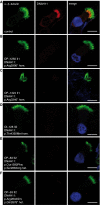
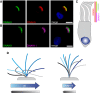
Primary ciliary dyskinesia (PCD) is a recessively inherited disease that leads to chronic respiratory disorders owing to impaired mucociliary clearance. Conventional transmission electron microscopy (TEM) is a diagnostic standard to identify ultrastructural defects in respiratory cilia but is not useful in approximately 30% of PCD cases, which have normal ciliary ultrastructure. DNAH11 mutations are a common cause of PCD with normal ciliary ultrastructure and hyperkinetic ciliary beating, but its pathophysiology remains poorly understood. We therefore characterized DNAH11 in human respiratory cilia by immunofluorescence microscopy (IFM) in the context of PCD. We used whole-exome and targeted next-generation sequence analysis as well as Sanger sequencing to identify and confirm eight novel loss-of-function DNAH11 mutations. We designed and validated a monoclonal antibody specific to DNAH11 and performed high-resolution IFM of both control and PCD-affected human respiratory cells, as well as samples from green fluorescent protein (GFP)-left-right dynein mice, to determine the ciliary localization of DNAH11. IFM analysis demonstrated native DNAH11 localization in only the proximal region of wild-type human respiratory cilia and loss of DNAH11 in individuals with PCD with certain loss-of-function DNAH11 mutations. GFP-left-right dynein mice confirmed proximal DNAH11 localization in tracheal cilia. DNAH11 retained proximal localization in respiratory cilia of individuals with PCD with distinct ultrastructural defects, such as the absence of outer dynein arms (ODAs). TEM tomography detected a partial reduction of ODAs in DNAH11-deficient cilia. DNAH11 mutations result in a subtle ODA defect in only the proximal region of respiratory cilia, which is detectable by IFM and TEM tomography.
Keywords: immunofluorescence microscopy; left–right dynein; normal ciliary ultrastructure; primary ciliary dyskinesia; transmission electron microscopy.
Figures

Similar articles
-
Recessive DNAH9 Loss-of-Function Mutations Cause Laterality Defects and Subtle Respiratory Ciliary-Beating Defects.Am J Hum Genet. 2018 Dec 6;103(6):995-1008. doi: 10.1016/j.ajhg.2018.10.020. Epub 2018 Nov 21. Am J Hum Genet. 2018. PMID: 30471718 Free PMC article.
-
Primary ciliary dyskinesia associated with normal axoneme ultrastructure is caused by DNAH11 mutations.Hum Mutat. 2008 Feb;29(2):289-98. doi: 10.1002/humu.20656. Hum Mutat. 2008. PMID: 18022865
-
Mutations of DNAH11 in patients with primary ciliary dyskinesia with normal ciliary ultrastructure.Thorax. 2012 May;67(5):433-41. doi: 10.1136/thoraxjnl-2011-200301. Epub 2011 Dec 18. Thorax. 2012. PMID: 22184204 Free PMC article.
-
Value of transmission electron microscopy for primary ciliary dyskinesia diagnosis in the era of molecular medicine: Genetic defects with normal and non-diagnostic ciliary ultrastructure.Ultrastruct Pathol. 2017 Nov-Dec;41(6):373-385. doi: 10.1080/01913123.2017.1362088. Epub 2017 Sep 15. Ultrastruct Pathol. 2017. PMID: 28915070 Free PMC article. Review.
-
[Cilia ultrastructural and gene variation of primary ciliary dyskinesia: report of three cases and literatures review].Zhonghua Er Ke Za Zhi. 2018 Feb 2;56(2):134-137. doi: 10.3760/cma.j.issn.0578-1310.2018.02.012. Zhonghua Er Ke Za Zhi. 2018. PMID: 29429202 Review. Chinese.
Cited by
-
Recessive DNAH9 Loss-of-Function Mutations Cause Laterality Defects and Subtle Respiratory Ciliary-Beating Defects.Am J Hum Genet. 2018 Dec 6;103(6):995-1008. doi: 10.1016/j.ajhg.2018.10.020. Epub 2018 Nov 21. Am J Hum Genet. 2018. PMID: 30471718 Free PMC article.
-
Empowering limited-resource countries: collaborating with expert centers for diagnosis of primary ciliary dyskinesia.Front Mol Biosci. 2025 Mar 20;12:1547152. doi: 10.3389/fmolb.2025.1547152. eCollection 2025. Front Mol Biosci. 2025. PMID: 40182617 Free PMC article.
-
Accuracy of Immunofluorescence in the Diagnosis of Primary Ciliary Dyskinesia.Am J Respir Crit Care Med. 2017 Jul 1;196(1):94-101. doi: 10.1164/rccm.201607-1351OC. Am J Respir Crit Care Med. 2017. PMID: 28199173 Free PMC article.
-
European Respiratory Society guidelines for the diagnosis of primary ciliary dyskinesia.Eur Respir J. 2017 Jan 4;49(1):1601090. doi: 10.1183/13993003.01090-2016. Print 2017 Jan. Eur Respir J. 2017. PMID: 27836958 Free PMC article.
-
Primary Ciliary Dyskinesia (PCD): A genetic disorder of motile cilia.Transl Sci Rare Dis. 2019;4(1-2):51-75. doi: 10.3233/TRD-190036. Epub 2019 Jul 4. Transl Sci Rare Dis. 2019. PMID: 31572664 Free PMC article. No abstract available.
References
-
- Fliegauf M, Benzing T, Omran H. When cilia go bad: cilia defects and ciliopathies. Nat Rev Mol Cell Biol. 2007;8:880–893. - PubMed
-
- Olbrich H, Häffner K, Kispert A, Völkel A, Volz A, Sasmaz G, Reinhardt R, Hennig S, Lehrach H, Konietzko N, et al. Mutations in DNAH5 cause primary ciliary dyskinesia and randomization of left–right asymmetry. Nat Genet. 2002;30:143–144. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

