Global hypermethylation in fetal cortex of Down syndrome due to DNMT3L overexpression
- PMID: 26911678
- PMCID: PMC4986328
- DOI: 10.1093/hmg/ddw043
Global hypermethylation in fetal cortex of Down syndrome due to DNMT3L overexpression
Abstract
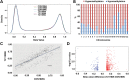
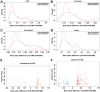
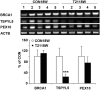
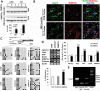
Down syndrome (DS) is caused by a triplication of chromosome 21 (HSA21). Increased oxidative stress, decreased neurogenesis and synaptic dysfunction from HSA21 gene overexpression are thought to cause mental retardation, dementia and seizure in this disorder. Recent epigenetic studies have raised the possibility that DNA methylation has significant effects on DS neurodevelopment. Here, we performed methylome profiling in normal and DS fetal cortices and observed a significant hypermethylation in ∼4% of probes in the DS samples compared with age-matched normals. The probes with differential methylation were distributed across all chromosomes, with no enrichment on HSA21. Functional annotation and pathway analyses showed that genes in the ubiquitination pathway were significantly altered, including: BRCA1, TSPYL5 and PEX10 HSA21 located DNMT3L was overexpressed in DS neuroprogenitors, and this overexpression increased the promoter methylation of TSPYL5 potentially through DNMT3B, and decreased its mRNA expression. DNMT3L overexpression also increased mRNA levels for TP53 and APP, effectors of TSPYL5 Furthermore, DNMT3L overexpression increased APP and PSD95 expression in differentiating neurons, whereas DNMT3LshRNA could partially rescue the APP and PSD95 up-regulation in DS cells. These results provide some of the first mechanistic insights into causes for epigenetic changes in DS, leading to modification of genes relevant for the DS neural endophenotype.
© The Author 2016. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures




References
-
- Esposito G., Imitola J., Lu J., De Filippis D., Scuderi C., Ganesh V.S., Folkerth R., Hecht J., Shin S., Iuvone T. et al. (2008) Genomic and functional profiling of human Down syndrome neural progenitors implicates S100B and aquaporin 4 in cell injury. Hum. Mol. Genet., 17, 440–457. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

