MicroRNAs as novel therapeutic targets to treat kidney injury and fibrosis
- PMID: 26911854
- PMCID: PMC5002060
- DOI: 10.1152/ajprenal.00523.2015
MicroRNAs as novel therapeutic targets to treat kidney injury and fibrosis
Abstract
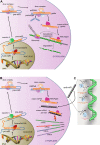
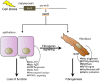
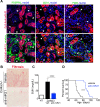
MicroRNAs (miRs), a class of small noncoding RNAs that act as post-transcriptional regulators of gene expression, have attracted increasing attention as critical regulators of organogenesis, cancer, and disease. Interest has been spurred by development of a novel class of synthetic RNA oligonucleotides with excellent drug-like properties that hybridize to a specific miR, preventing its action. In kidney disease, a small number of miRs are dysregulated. These overlap with regulated miRs in nephrogenesis and kidney cancers. Several dysregulated miRs have been identified in fibrotic diseases of other organs, representing a "fibrotic signature," and some of these fibrotic miRs contribute remarkably to the pathogenesis of kidney disease. Chronic kidney disease, affecting ∼10% of the population, leads to kidney failure, with few treatment options. Here, we will explore the pathological mechanism of miR-21, whose pre-eminent role in amplifying kidney disease and fibrosis by suppressing mitochondrial biogenesis and function is established. Evolving roles for miR-214, -199, -200, -155, -29, -223, and -126 in kidney disease will be discussed, and we will demonstrate how studying functions of distinct miRs has led to new mechanistic insights for kidney disease progression. Finally, the utility of anti-miR oligonucleotides as potential novel therapeutics to treat chronic disease will be highlighted.
Keywords: Dicer; angiogenesis; chronic kidney disease; fatty acid oxidation; macrophage activation; microRNAs; mitochondria.
Copyright © 2016 the American Physiological Society.
Figures


References
-
- Azzouzi el H, Leptidis S, Dirkx E, Hoeks J, van Bree B, Brand K, McClellan EA, Poels E, Sluimer JC, van den Hoogenhof MMG, Armand AS, Yin X, Langley S, Bourajjaj M, Olieslagers S, Krishnan J, Vooijs M, Kurihara H, Stubbs A, Pinto YM, Krek W, Mayr M, da Costa Martins PA, Schrauwen P, De Windt LJ. The hypoxia-inducible microRNA cluster miR-199a∼214 targets myocardial PPARδ and impairs mitochondrial fatty acid oxidation. Cell Metab 18: 341–354, 2013. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

