Chlamydia trachomatis from Australian Aboriginal people with trachoma are polyphyletic composed of multiple distinctive lineages
- PMID: 26912299
- PMCID: PMC4773424
- DOI: 10.1038/ncomms10688
Chlamydia trachomatis from Australian Aboriginal people with trachoma are polyphyletic composed of multiple distinctive lineages
Abstract
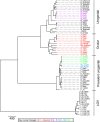
Chlamydia trachomatis causes sexually transmitted infections and the blinding disease trachoma. Current data on C. trachomatis phylogeny show that there is only a single trachoma-causing clade, which is distinct from the lineages causing urogenital tract (UGT) and lymphogranuloma venerum diseases. Here we report the whole-genome sequences of ocular C. trachomatis isolates obtained from young children with clinical signs of trachoma in a trachoma endemic region of northern Australia. The isolates form two lineages that fall outside the classical trachoma lineage, instead being placed within UGT clades of the C. trachomatis phylogenetic tree. The Australian trachoma isolates appear to be recombinants with UGT C. trachomatis genome backbones, in which loci that encode immunodominant surface proteins (ompA and pmpEFGH) have been replaced by those characteristic of classical ocular isolates. This suggests that ocular tropism and association with trachoma are functionally associated with some sequence variants of ompA and pmpEFGH.
Figures





References
-
- Mariotti S. P., Pascolini D. & Rose-Nussbaumer J. Trachoma: global magnitude of a preventable cause of blindness. Br. J. Ophthalmol. 93, 563–568 (2009). - PubMed
-
- WHO. Prevalence and Incidence of Selected Sexually Transmitted Infections World Health Organization (2011).
-
- Taylor H. R. (ed.). Trachoma: A Blinding Scourge from the Bronze Age to the Twenty-First Century Centre for Eye Research Australia (2008).
-
- Stephens R. S., Tam M. R., Kuo C. C. & Nowinski R. C. Monoclonal antibodies to Chlamydia trachomatis: antibody specificities and antigen characterization. J. Immunol. 128, 1083–1089 (1982). - PubMed
-
- Wang S. P., Kuo C. C., Barnes R. C., Stephens R. S. & Grayston J. T. Immunotyping of Chlamydia trachomatis with monoclonal antibodies. J. Infect. Dis. 152, 791–800 (1985). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

