Isolation of potent neutralizing antibodies from a survivor of the 2014 Ebola virus outbreak
- PMID: 26912366
- PMCID: PMC4900763
- DOI: 10.1126/science.aad5788
Isolation of potent neutralizing antibodies from a survivor of the 2014 Ebola virus outbreak
Abstract
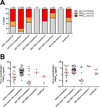
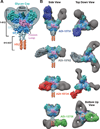
Antibodies targeting the Ebola virus surface glycoprotein (EBOV GP) are implicated in protection against lethal disease, but the characteristics of the human antibody response to EBOV GP remain poorly understood. We isolated and characterized 349 GP-specific monoclonal antibodies (mAbs) from the peripheral B cells of a convalescent donor who survived the 2014 EBOV Zaire outbreak. Remarkably, 77% of the mAbs neutralize live EBOV, and several mAbs exhibit unprecedented potency. Structures of selected mAbs in complex with GP reveal a site of vulnerability located in the GP stalk region proximal to the viral membrane. Neutralizing antibodies targeting this site show potent therapeutic efficacy against lethal EBOV challenge in mice. The results provide a framework for the design of new EBOV vaccine candidates and immunotherapies.
Copyright © 2016, American Association for the Advancement of Science.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

