Functional Dissection of an Alternatively Spliced Herpesvirus Gene by Splice Site Mutagenesis
- PMID: 26912612
- PMCID: PMC4836318
- DOI: 10.1128/JVI.02987-15
Functional Dissection of an Alternatively Spliced Herpesvirus Gene by Splice Site Mutagenesis
Abstract
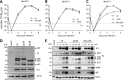
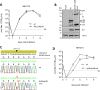
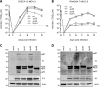
Herpesviruses have large and complex DNA genomes. The largest among the herpesviruses, those of the cytomegaloviruses, include over 170 genes. Although most herpesvirus gene products are expressed from unspliced transcripts, a substantial number of viral transcripts are spliced. Some viral transcripts are subject to alternative splicing, which leads to the expression of several proteins from a single gene. Functional analysis of individual proteins derived from an alternatively spliced gene is difficult, as deletion and nonsense mutagenesis, both common methods used in the generation of viral gene knockout mutants, affect several or all gene products at the same time. Here, we show that individual gene products of an alternatively spliced herpesvirus gene can be inactivated selectively by mutagenesis of the splice donor or acceptor site and by intron deletion or substitution mutagenesis. We used this strategy to dissect the essential M112/113 gene of murine cytomegalovirus (MCMV), which encodes the MCMV Early 1 (E1) proteins. The expression of each of the four E1 protein isoforms was inactivated individually, and the requirement for each isoform in MCMV replication was analyzed in fibroblasts, endothelial cells, and macrophages. We show that the E1 p87 isoform, but not the p33, p36, and p38 isoforms, is essential for viral replication in cell culture. Moreover, the presence of one of the two medium-size isoforms (p36 or p38) and the presence of intron 1, but not its specific sequence, are required for viral replication. This study demonstrates the usefulness of splice site mutagenesis for the functional analysis of alternatively spliced herpesvirus genes.
Importance: Herpesviruses include up to 170 genes in their DNA genomes. The functions of most viral gene products remain poorly defined. The construction of viral gene knockout mutants has thus been an important tool for functional analysis of viral proteins. However, this strategy is of limited use when viral gene transcripts are alternatively spliced, leading to the expression of several proteins from a single gene. In this study, we showed, as a proof of principle, that each protein product of an alternatively spliced gene can be eliminated individually by splice site mutagenesis. Mutant viruses lacking individual protein products displayed different phenotypes, demonstrating that the products of alternatively spliced genes have nonredundant functions.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures



Similar articles
-
Differential Requirement of Human Cytomegalovirus UL112-113 Protein Isoforms for Viral Replication.J Virol. 2017 Aug 10;91(17):e00254-17. doi: 10.1128/JVI.00254-17. Print 2017 Sep 1. J Virol. 2017. PMID: 28637762 Free PMC article.
-
The Canonical Immediate Early 3 Gene Product pIE611 of Mouse Cytomegalovirus Is Dispensable for Viral Replication but Mediates Transcriptional and Posttranscriptional Regulation of Viral Gene Products.J Virol. 2015 Aug;89(16):8590-8. doi: 10.1128/JVI.01234-15. Epub 2015 Jun 10. J Virol. 2015. PMID: 26063418 Free PMC article.
-
Identification and characterization of novel murine cytomegalovirus M112-113 (e1) gene products.Virology. 2002 Mar 1;294(1):199-208. doi: 10.1006/viro.2001.1311. Virology. 2002. PMID: 11886278
-
Genetic analyses of gene function and pathogenesis of murine cytomegalovirus by transposon-mediated mutagenesis.J Clin Virol. 2002 Aug;25 Suppl 2:S111-22. doi: 10.1016/s1386-6532(02)00096-3. J Clin Virol. 2002. PMID: 12361762 Review.
-
Herpesvirus genes: molecular basis of viral replication and pathogenicity.Nagoya J Med Sci. 1996 Dec;59(3-4):107-19. Nagoya J Med Sci. 1996. PMID: 9212636 Review.
Cited by
-
Prolonged activation of cytomegalovirus early gene e1-promoter exclusively in neurons during infection of the developing cerebrum.Acta Neuropathol Commun. 2021 Mar 9;9(1):39. doi: 10.1186/s40478-021-01139-0. Acta Neuropathol Commun. 2021. PMID: 33750455 Free PMC article.
-
Alternative splicing isoforms in health and disease.Pflugers Arch. 2018 Jul;470(7):995-1016. doi: 10.1007/s00424-018-2136-x. Epub 2018 Mar 13. Pflugers Arch. 2018. PMID: 29536164 Review.
-
Human cytomegalovirus forms phase-separated compartments at viral genomes to facilitate viral replication.Cell Rep. 2022 Mar 8;38(10):110469. doi: 10.1016/j.celrep.2022.110469. Cell Rep. 2022. PMID: 35263605 Free PMC article.
-
Long and Short Isoforms of the Human Cytomegalovirus UL138 Protein Silence IE Transcription and Promote Latency.J Virol. 2016 Sep 29;90(20):9483-94. doi: 10.1128/JVI.01547-16. Print 2016 Oct 15. J Virol. 2016. PMID: 27512069 Free PMC article.
-
Murine cytomegaloviruses m139 targets DDX3 to curtail interferon production and promote viral replication.PLoS Pathog. 2020 Oct 8;16(10):e1008546. doi: 10.1371/journal.ppat.1008546. eCollection 2020 Oct. PLoS Pathog. 2020. PMID: 33031466 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

