Quantification of Cooperativity in Heterodimer-DNA Binding Improves the Accuracy of Binding Specificity Models
- PMID: 26912662
- PMCID: PMC4858977
- DOI: 10.1074/jbc.M115.691154
Quantification of Cooperativity in Heterodimer-DNA Binding Improves the Accuracy of Binding Specificity Models
Abstract
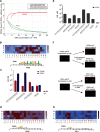
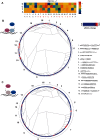
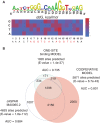
Many transcription factors (TFs) have the ability to cooperate on DNA elements as heterodimers. Despite the significance of TF heterodimerization for gene regulation, a quantitative understanding of cooperativity between various TF dimer partners and its impact on heterodimer DNA binding specificity models is still lacking. Here, we used a novel integrative approach, combining microfluidics-steered measurements of dimer-DNA assembly with mechanistic modeling of the implicated protein-protein-DNA interactions to quantitatively interrogate the cooperative DNA binding behavior of the adipogenic peroxisome proliferator-activated receptor γ (PPARγ):retinoid X receptor α (RXRα) heterodimer. Using the high throughput MITOMI (mechanically induced trapping of molecular interactions) platform, we derived equilibrium DNA binding data for PPARγ, RXRα, as well as the PPARγ:RXRα heterodimer to more than 300 target DNA sites and variants thereof. We then quantified cooperativity underlying heterodimer-DNA binding and derived an integrative heterodimer DNA binding constant. Using this cooperativity-inclusive constant, we were able to build a heterodimer-DNA binding specificity model that has superior predictive power than the one based on a regular one-site equilibrium. Our data further revealed that individual nucleotide substitutions within the target site affect the extent of cooperativity in PPARγ:RXRα-DNA binding. Our study therefore emphasizes the importance of assessing cooperativity when generating DNA binding specificity models for heterodimers.
Keywords: DNA-binding protein; computational biology; cooperativity; heterodimer-DNA interactions; specificity models; transcription; transcription regulation.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures




References
-
- Davidson E. H., and Erwin D. H. (2006) Gene regulatory networks and the evolution of animal body plans. Science 311, 796–800 - PubMed
-
- Deplancke B. (2009) Experimental advances in the characterization of metazoan gene regulatory networks. Brief. Funct. Genomic Proteomic 8, 12–27 - PubMed
-
- Badis G., Berger M. F., Philippakis A. A., Talukder S., Gehrke A. R., Jaeger S. A., Chan E. T., Metzler G., Vedenko A., Chen X., Kuznetsov H., Wang C. F., Coburn D., Newburger D. E., Morris Q., et al. (2009) Diversity and complexity in DNA recognition by transcription factors. Science 324, 1720–1723 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

