Pathogenic CD4 T cells in type 1 diabetes recognize epitopes formed by peptide fusion
- PMID: 26912858
- PMCID: PMC4884646
- DOI: 10.1126/science.aad2791
Pathogenic CD4 T cells in type 1 diabetes recognize epitopes formed by peptide fusion
Abstract
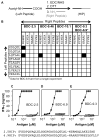
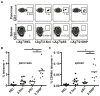
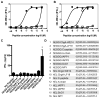
T cell-mediated destruction of insulin-producing β cells in the pancreas causes type 1 diabetes (T1D). CD4 T cell responses play a central role in β cell destruction, but the identity of the epitopes recognized by pathogenic CD4 T cells remains unknown. We found that diabetes-inducing CD4 T cell clones isolated from nonobese diabetic mice recognize epitopes formed by covalent cross-linking of proinsulin peptides to other peptides present in β cell secretory granules. These hybrid insulin peptides (HIPs) are antigenic for CD4 T cells and can be detected by mass spectrometry in β cells. CD4 T cells from the residual pancreatic islets of two organ donors who had T1D also recognize HIPs. Autoreactive T cells targeting hybrid peptides may explain how immune tolerance is broken in T1D.
Copyright © 2016, American Association for the Advancement of Science.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
- 1R01DK081166/DK/NIDDK NIH HHS/United States
- 1K01DK094941/DK/NIDDK NIH HHS/United States
- P60 DK020593/DK/NIDDK NIH HHS/United States
- UC4 DK104211/DK/NIDDK NIH HHS/United States
- DK104211/DK/NIDDK NIH HHS/United States
- R01 DK081166/DK/NIDDK NIH HHS/United States
- P30 DK032520/DK/NIDDK NIH HHS/United States
- U01 DK072473/DK/NIDDK NIH HHS/United States
- U01 DK089572/DK/NIDDK NIH HHS/United States
- I01 BX000666/BX/BLRD VA/United States
- P30 DK020593/DK/NIDDK NIH HHS/United States
- K01 DK094941/DK/NIDDK NIH HHS/United States
- 5U01DK89572/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

