BioShaDock: a community driven bioinformatics shared Docker-based tools registry
- PMID: 26913191
- PMCID: PMC4743153
- DOI: 10.12688/f1000research.7536.1
BioShaDock: a community driven bioinformatics shared Docker-based tools registry
Abstract
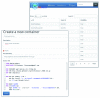
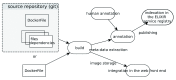
Linux container technologies, as represented by Docker, provide an alternative to complex and time-consuming installation processes needed for scientific software. The ease of deployment and the process isolation they enable, as well as the reproducibility they permit across environments and versions, are among the qualities that make them interesting candidates for the construction of bioinformatic infrastructures, at any scale from single workstations to high throughput computing architectures. The Docker Hub is a public registry which can be used to distribute bioinformatic software as Docker images. However, its lack of curation and its genericity make it difficult for a bioinformatics user to find the most appropriate images needed. BioShaDock is a bioinformatics-focused Docker registry, which provides a local and fully controlled environment to build and publish bioinformatic software as portable Docker images. It provides a number of improvements over the base Docker registry on authentication and permissions management, that enable its integration in existing bioinformatic infrastructures such as computing platforms. The metadata associated with the registered images are domain-centric, including for instance concepts defined in the EDAM ontology, a shared and structured vocabulary of commonly used terms in bioinformatics. The registry also includes user defined tags to facilitate its discovery, as well as a link to the tool description in the ELIXIR registry if it already exists. If it does not, the BioShaDock registry will synchronize with the registry to create a new description in the Elixir registry, based on the BioShaDock entry metadata. This link will help users get more information on the tool such as its EDAM operations, input and output types. This allows integration with the ELIXIR Tools and Data Services Registry, thus providing the appropriate visibility of such images to the bioinformatics community.
Keywords: bioinformatics; community driven registry; container; deployment; docker; interoperability; maintainability.
Conflict of interest statement
Figures

References
-
- Connor BO, Kartashov A, Yuen D, et al. : ELIXIR Tools and Data Services Registry.2015. Reference Source
LinkOut - more resources
Full Text Sources
Other Literature Sources

