Increased expression of long noncoding RNA TUG1 predicts a poor prognosis of gastric cancer and regulates cell proliferation by epigenetically silencing of p57
- PMID: 26913601
- PMCID: PMC4849144
- DOI: 10.1038/cddis.2015.356
Increased expression of long noncoding RNA TUG1 predicts a poor prognosis of gastric cancer and regulates cell proliferation by epigenetically silencing of p57
Abstract
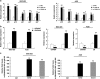
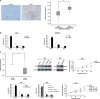
Recent evidence highlights long noncoding RNAs (lncRNAs) as crucial regulators of cancer biology that contribute to tumorigenesis. LncRNA TUG1 was initially detected in a genomic screen for genes upregulated in response to taurine treatment in developing mouse retinal cells. Our previous study showed that TUG1 could affect cell proliferation through epigenetically regulating HOXB7 in human non-small cell lung cancer. However, the clinical significance and potential role of TUG1 in GC remains unclear. In this study, we found that TUG1 is significantly increased and is correlated with outcomes in gastric cancer (GC). Further experiments revealed that knockdown of TUG1 repressed GC proliferation both in vitro and in vivo. Mechanistic investigations showed that TUG1 has a key role in G0/G1 arrest. We further demonstrated that TUG1 was associated with PRC2 and that this association was required for epigenetic repression of cyclin-dependent protein kinase inhibitors, including p15, p16, p21, p27 and p57, thus contributing to the regulation of GC cell cycle and proliferation. Together, our results suggest that TUG1, as a regulator of proliferation, may serve as a candidate prognostic biomarker and target for new therapies in human GC.
Figures




References
-
- Coupland VH, Lagergren J, Luchtenborg M, Jack RH, Allum W, Holmberg L et al. Hospital volume, proportion resected and mortality from oesophageal and gastric cancer: a population-based study in England, 2004-2008. Gut 2013; 62: 961–966. - PubMed
-
- Amaral PP, Dinger ME, Mercer TR, Mattick JS. The eukaryotic genome as an RNA machine. Science 2008; 319: 1787–1789. - PubMed
-
- Nagano T, Fraser P. No-nonsense functions for long noncoding RNAs. Cell 2011; 145: 178–181. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

