Incorporating a Genetic Risk Score Into Coronary Heart Disease Risk Estimates: Effect on Low-Density Lipoprotein Cholesterol Levels (the MI-GENES Clinical Trial)
- PMID: 26915630
- PMCID: PMC4803581
- DOI: 10.1161/CIRCULATIONAHA.115.020109
Incorporating a Genetic Risk Score Into Coronary Heart Disease Risk Estimates: Effect on Low-Density Lipoprotein Cholesterol Levels (the MI-GENES Clinical Trial)
Abstract
Background: Whether knowledge of genetic risk for coronary heart disease (CHD) affects health-related outcomes is unknown. We investigated whether incorporating a genetic risk score (GRS) in CHD risk estimates lowers low-density lipoprotein cholesterol (LDL-C) levels.
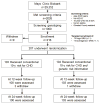
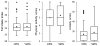
Methods and results: Participants (n=203, 45-65 years of age, at intermediate risk for CHD, and not on statins) were randomly assigned to receive their 10-year probability of CHD based either on a conventional risk score (CRS) or CRS + GRS ((+)GRS). Participants in the (+)GRS group were stratified as having high or average/low GRS. Risk was disclosed by a genetic counselor followed by shared decision making regarding statin therapy with a physician. We compared the primary end point of LDL-C levels at 6 months and assessed whether any differences were attributable to changes in dietary fat intake, physical activity levels, or statin use. Participants (mean age, 59.4±5 years; 48% men; mean 10-year CHD risk, 8.5±4.1%) were allocated to receive either CRS (n=100) or (+)GRS (n=103). At the end of the study period, the (+)GRS group had a lower LDL-C than the CRS group (96.5±32.7 versus 105.9±33.3 mg/dL; P=0.04). Participants with high GRS had lower LDL-C levels (92.3±32.9 mg/dL) than CRS participants (P=0.02) but not participants with low GRS (100.9±32.2 mg/dL; P=0.18). Statins were initiated more often in the (+)GRS group than in the CRS group (39% versus 22%, P<0.01). No significant differences in dietary fat intake and physical activity levels were noted.
Conclusions: Disclosure of CHD risk estimates that incorporated genetic risk information led to lower LDL-C levels than disclosure of CHD risk based on conventional risk factors alone.
Clinical trial registration: URL: http://www.clinicaltrials.gov. Unique identifier: NCT01936675.
Keywords: coronary disease; genetic risk disclosure; genetic risk score; genomics; hydroxymethylglutaryl-CoA reductase inhibitors; polymorphism, single-nucleotide; prevention & control; randomized clinical trials.
© 2016 American Heart Association, Inc.
Figures
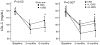
Comment in
-
Reducing Cardiovascular Risk Using Genomic Information in the Era of Precision Medicine.Circulation. 2016 Mar 22;133(12):1155-9. doi: 10.1161/CIRCULATIONAHA.116.021765. Epub 2016 Feb 25. Circulation. 2016. PMID: 26915631 Free PMC article. No abstract available.
References
-
- Manolio TA. Bringing genome-wide association findings into clinical use. Nat Rev Genet. 2013;14:549–558. - PubMed
-
- The Coronary Artery Disease (C4D) Genetics Consortium. A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat Genet. 2011;43:339–344. - PubMed
-
- Deloukas P, Kanoni S, Willenborg C, Farrall M, Assimes TL, Thompson JR, Ingelsson E, Saleheen D, Erdmann J, Goldstein BA, Stirrups K, Konig IR, Cazier JB, Johansson A, Hall AS, Lee JY, Willer CJ, Chambers JC, Esko T, Folkersen L, Goel A, Grundberg E, Havulinna AS, Ho WK, Hopewell JC, Eriksson N, Kleber ME, Kristiansson K, Lundmark P, Lyytikainen LP, Rafelt S, Shungin D, Strawbridge RJ, Thorleifsson G, Tikkanen E, Van Zuydam N, Voight BF, Waite LL, Zhang W, Ziegler A, Absher D, Altshuler D, Balmforth AJ, Barroso I, Braund PS, Burgdorf C, Claudi-Boehm S, Cox D, Dimitriou M, Do R, Doney AS, El Mokhtari N, Eriksson P, Fischer K, Fontanillas P, Franco-Cereceda A, Gigante B, Groop L, Gustafsson S, Hager J, Hallmans G, Han BG, Hunt SE, Kang HM, Illig T, Kessler T, Knowles JW, Kolovou G, Kuusisto J, Langenberg C, Langford C, Leander K, Lokki ML, Lundmark A, McCarthy MI, Meisinger C, Melander O, Mihailov E, Maouche S, Morris AD, Muller-Nurasyid M, Nikus K, Peden JF, Rayner NW, Rasheed A, Rosinger S, Rubin D, Rumpf MP, Schafer A, Sivananthan M, Song C, Stewart AF, Tan ST, Thorgeirsson G, van der Schoot CE, Wagner PJ, Wells GA, Wild PS, Yang TP, Amouyel P, Arveiler D, Basart H, Boehnke M, Boerwinkle E, Brambilla P, Cambien F, Cupples AL, de Faire U, Dehghan A, Diemert P, Epstein SE, Evans A, Ferrario MM, Ferrieres J, Gauguier D, Go AS, Goodall AH, Gudnason V, Hazen SL, Holm H, Iribarren C, Jang Y, Kahonen M, Kee F, Kim HS, Klopp N, Koenig W, Kratzer W, Kuulasmaa K, Laakso M, Laaksonen R, Lind L, Ouwehand WH, Parish S, Park JE, Pedersen NL, Peters A, Quertermous T, Rader DJ, Salomaa V, Schadt E, Shah SH, Sinisalo J, Stark K, Stefansson K, Tregouet DA, Virtamo J, Wallentin L, Wareham N, Zimmermann ME, Nieminen MS, Hengstenberg C, Sandhu MS, Pastinen T, Syvanen AC, Hovingh GK, Dedoussis G, Franks PW, Lehtimaki T, Metspalu A, Zalloua PA, Siegbahn A, Schreiber S, Ripatti S, Blankenberg SS, Perola M, Clarke R, Boehm BO, O’Donnell C, Reilly MP, Marz W, Collins R, Kathiresan S, Hamsten A, Kooner JS, Thorsteinsdottir U, Danesh J, Palmer CN, Roberts R, Watkins H, Schunkert H, Samani NJ. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet. 2013;45:25–33. - PMC - PubMed
-
- Thanassoulis G, Peloso GM, Pencina MJ, Hoffmann U, Fox CS, Cupples LA, Levy D, D’Agostino RB, Hwang SJ, O’Donnell CJ. A genetic risk score is associated with incident cardiovascular disease and coronary artery calcium: the Framingham Heart Study. Circ Cardiovasc Genet. 2012;5:113–121. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

