Culture-independent method for identification of microbial enzyme-encoding genes by activity-based single-cell sequencing using a water-in-oil microdroplet platform
- PMID: 26915788
- PMCID: PMC4768102
- DOI: 10.1038/srep22259
Culture-independent method for identification of microbial enzyme-encoding genes by activity-based single-cell sequencing using a water-in-oil microdroplet platform
Abstract
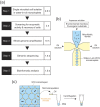
Environmental microbes are a great source of industrially valuable enzymes with potent and unique catalytic activities. Unfortunately, the majority of microbes remain unculturable and thus are not accessible by culture-based methods. Recently, culture-independent metagenomic approaches have been successfully applied, opening access to untapped genetic resources. Here we present a methodological approach for the identification of genes that encode metabolically active enzymes in environmental microbes in a culture-independent manner. Our method is based on activity-based single-cell sequencing, which focuses on microbial cells showing specific enzymatic activities. First, at the single-cell level, environmental microbes were encapsulated in water-in-oil microdroplets with a fluorogenic substrate for the target enzyme to screen for microdroplets that contain microbially active cells. Second, the microbial cells were recovered and subjected to whole genome amplification. Finally, the amplified genomes were sequenced to identify the genes encoding target enzymes. Employing this method, we successfully identified 14 novel β-glucosidase genes from uncultured bacterial cells in marine samples. Our method contributes to the screening and identification of genes encoding industrially valuable enzymes.
Figures

References
-
- Sanchez S. & Demain A. L. Enzymes and bioconversions of industrial, pharmaceutical, and biotechnological significance. Org. Process. Res. Dev. 15, 224–230 (2011).
-
- Riesenfeld C. S., Schloss P. D. & Handelsman J. Metagenomics: genomic analysis of microbial communities. Annu. Rev. Genet. 38, 525–552 (2004). - PubMed
-
- Lorenz P. & Eck J. Metagenomics and industrial applications. Nat. Rev. Microbiol. 3, 510−516 (2005). - PubMed
-
- Uchiyama T. & Miyazaki K. Functional metagenomics for enzyme discovery: challenges to efficient screening. Curr. Opin. Biotechnol. 20, 616–622 (2009). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

