Molecular epidemiology and evolutionary histories of human coronavirus OC43 and HKU1 among patients with upper respiratory tract infections in Kuala Lumpur, Malaysia
- PMID: 26916286
- PMCID: PMC4766700
- DOI: 10.1186/s12985-016-0488-4
Molecular epidemiology and evolutionary histories of human coronavirus OC43 and HKU1 among patients with upper respiratory tract infections in Kuala Lumpur, Malaysia
Abstract
Background: Despite the worldwide circulation of human coronavirus OC43 (HCoV-OC43) and HKU1 (HCoV-HKU1), data on their molecular epidemiology and evolutionary dynamics in the tropical Southeast Asia region is lacking.
Methods: The study aimed to investigate the genetic diversity, temporal distribution, population history and clinical symptoms of betacoronavirus infections in Kuala Lumpur, Malaysia between 2012 and 2013. A total of 2,060 adults presented with acute respiratory symptoms were screened for the presence of betacoronaviruses using multiplex PCR. The spike glycoprotein, nucleocapsid and 1a genes were sequenced for phylogenetic reconstruction and Bayesian coalescent inference.
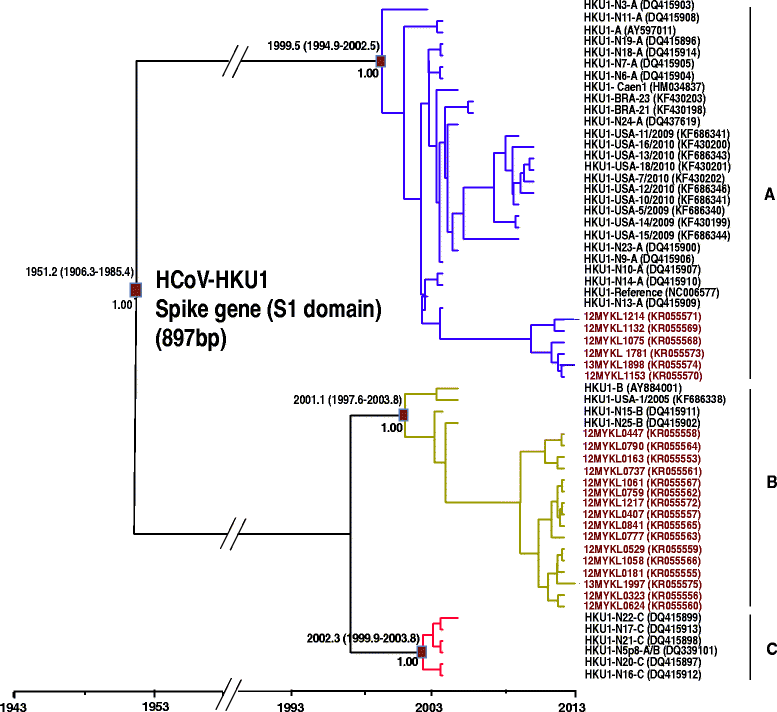
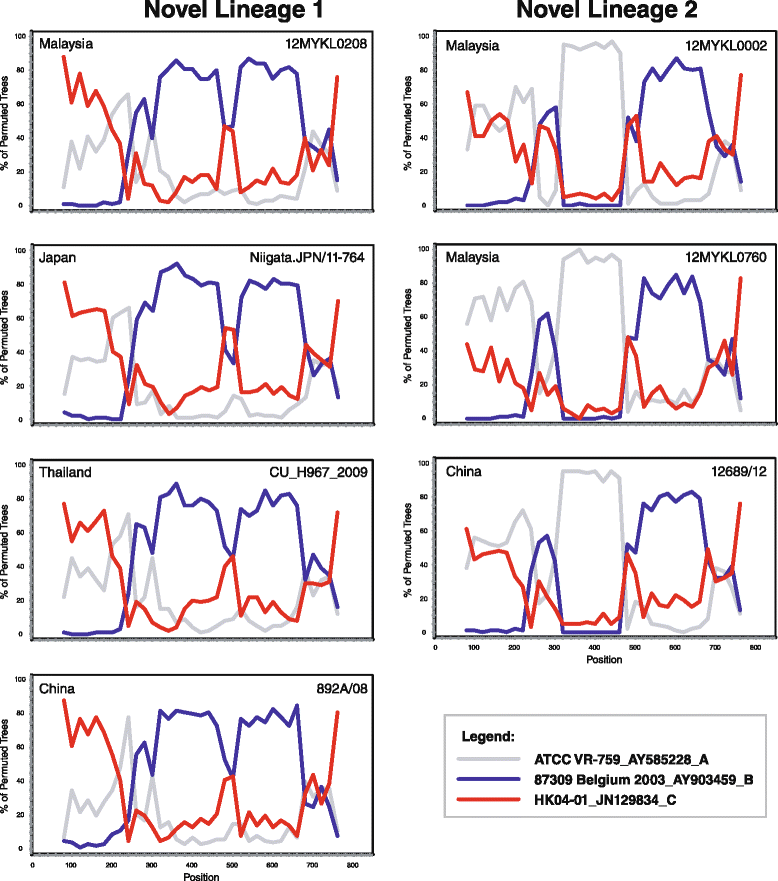
Results: A total of 48/2060 (2.4 %) specimens were tested positive for HCoV-OC43 (1.3 %) and HCoV-HKU1 (1.1 %). Both HCoV-OC43 and HCoV-HKU1 were co-circulating throughout the year, with the lowest detection rates reported in the October-January period. Phylogenetic analysis of the spike gene showed that the majority of HCoV-OC43 isolates were grouped into two previously undefined genotypes, provisionally assigned as novel lineage 1 and novel lineage 2. Sign of natural recombination was observed in these potentially novel lineages. Location mapping showed that the novel lineage 1 is currently circulating in Malaysia, Thailand, Japan and China, while novel lineage 2 can be found in Malaysia and China. Molecular dating showed the origin of HCoV-OC43 around late 1950s, before it diverged into genotypes A (1960s), B (1990s), and other genotypes (2000s). Phylogenetic analysis revealed that 27.3 % of the HCoV-HKU1 strains belong to genotype A while 72.7 % belongs to genotype B. The tree root of HCoV-HKU1 was similar to that of HCoV-OC43, with the tMRCA of genotypes A and B estimated around the 1990s and 2000s, respectively. Correlation of HCoV-OC43 and HCoV-HKU1 with the severity of respiratory symptoms was not observed.
Conclusions: The present study reported the molecular complexity and evolutionary dynamics of human betacoronaviruses among adults with acute respiratory symptoms in a tropical country. Two novel HCoV-OC43 genetic lineages were identified, warranting further investigation on their genotypic and phenotypic characteristics.
Figures


References
-
- Vijgen L, Keyaerts E, Moes E, Maes P, Duson G, Van Ranst M. Development of one-step, real-time, quantitative reverse transcriptase PCR assays for absolute quantitation of human coronaviruses OC43 and 229E. J Clin Microbiol. 2005;43(11):5452–6. doi: 10.1128/JCM.43.11.5452-5456.2005. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

