Retention of hexanucleotide repeat-containing intron in C9orf72 mRNA: implications for the pathogenesis of ALS/FTD
- PMID: 26916632
- PMCID: PMC4766718
- DOI: 10.1186/s40478-016-0289-4
Retention of hexanucleotide repeat-containing intron in C9orf72 mRNA: implications for the pathogenesis of ALS/FTD
Abstract
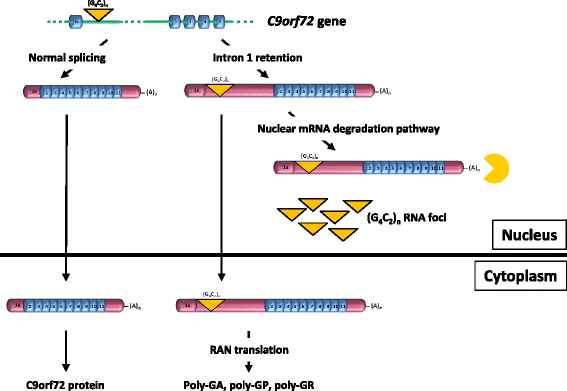
Introduction: The most common forms of amyotrophic lateral sclerosis and frontotemporal dementia are caused by a large GGGGCC repeat expansion in the first intron of the C9orf72 gene. The repeat-containing intron should be degraded after being spliced out, however GGGGCC repeat-containing RNA species either accumulate in nuclear foci or are exported to the cytoplasm where they are translated into potentially toxic dipeptide repeat proteins by repeat-associated non-AUG-initiated (RAN) translation.
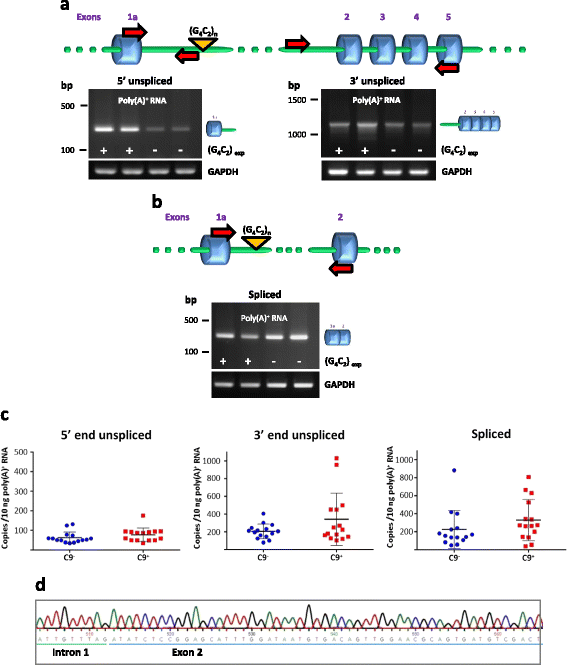
Results: In order to determine the mechanisms of repeat-containing intron misprocessing, we have analyzed C9orf72 transcripts in lymphoblasts from C9orf72 expansion carriers (n = 15) and control individuals (n = 15). We have identified polyadenylated C9orf72 RNA species retaining the repeat-containing intron and in which downstream exons are spliced correctly resulting in a C9orf72 mRNA with an enlarged 5'-UTR containing the GGGGCC repeats. Intron-retaining transcripts are produced from both wild-type and mutant alleles. Intron-retaining C9orf72 transcripts were also detected in brain with a 2.7 fold increase measured in the frontal cortex from heterozygous expansion carriers (n = 11) compared to controls (n = 10). The level of intron-retaining transcripts was increased 5.9 fold in a case homozygous for the expansion. We also show that a large proportion of intron 1-retaining C9orf72 transcripts accumulate in the nucleus.
Conclusions: Retention of the repeat-containing intron in mature C9orf72 mRNA can potentially explain nuclear foci formation as well as nuclear export of GGGGCC repeat RNA and suggests that the misprocessing of C9orf72 transcripts initiates the pathogenic process caused by C9orf72 hexanucleotide repeat expansions as well as provides the basis for novel therapeutic strategies.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
- GALLO/APR14/827-791/MNDA_/Motor Neurone Disease Association/United Kingdom
- 089701/WT_/Wellcome Trust/United Kingdom
- SHAW/APR15/970-797/MNDA_/Motor Neurone Disease Association/United Kingdom
- G1100695/MRC_/Medical Research Council/United Kingdom
- MR/L016397/1/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

