Role of Flippases in Protein Glycosylation in the Endoplasmic Reticulum
- PMID: 26917968
- PMCID: PMC4762491
- DOI: 10.4137/LPI.S31784
Role of Flippases in Protein Glycosylation in the Endoplasmic Reticulum
Abstract
Glycosylation is essential to the synthesis, folding, and function of glycoproteins in eukaryotes. Proteins are co- and posttranslationally modified by a variety of glycans in the endoplasmic reticulum (ER); modifications include C- and O-mannosylation, N-glycosylation, and the addition of glycosylphosphatidylinositol membrane anchors. Protein glycosylation in the ER of eukaryotes involves enzymatic steps on both the cytosolic and lumenal surfaces of the ER membrane. The glycans are first assembled as precursor glycolipids, on the cytosolic surface of the ER, which are tethered to the membrane by attachment to a long-chain polyisoprenyl phosphate (dolichol) containing a reduced α-isoprene. The lipid-anchored building blocks then migrate transversely (flip) across the ER membrane to the lumenal surface, where final assembly of the glycan is completed. This strategy allows the cell to export high-energy biosynthetic intermediates as lipid-bound glycans, while constraining the glycosyl donors to the site of assembly on the membrane surface. This review focuses on the flippases that participate in protein glycosylation in the ER.
Keywords: congenital disorder of glycosylation; dolichol; flippase; glycosylation.
Figures

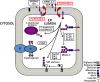

References
-
- Holthuis JC, Menon AK. Lipid landscapes and pipelines in membrane homeostasis. Nature. 2014;510(7503):48–57. - PubMed
-
- Pomorski T, Holthuis JC, Herrmann A, van Meer G. Tracking down lipid flippases and their biological functions. J Cell Sci. 2004;117(pt 6):805–813. - PubMed
-
- Panatala R, Hennrich H, Holthuis JC. Inner workings and biological impact of phospholipid flippases. J Cell Sci. 2015;128(11):2021–2032. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

