Current approaches to studying membrane organization
- PMID: 26918150
- PMCID: PMC4754029
- DOI: 10.12688/f1000research.6868.1
Current approaches to studying membrane organization
Abstract
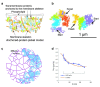
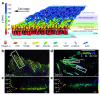
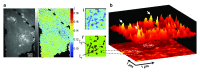
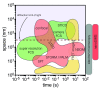
The local structure and composition of the outer membrane of an animal cell are important factors in the control of many membrane processes and mechanisms. These include signaling, sorting, and exo- and endocytic processes that are occurring all the time in a living cell. Paradoxically, not only are the local structure and composition of the membrane matters of much debate and discussion, the mechanisms that govern its genesis remain highly controversial. Here, we discuss a swathe of new technological advances that may be applied to understand the local structure and composition of the membrane of a living cell from the molecular scale to the scale of the whole membrane.
Keywords: electron microscopy; membrane; proteins.
Conflict of interest statement
No competing interests were disclosed.
Figures

Similar articles
-
Actomyosin dynamics drive local membrane component organization in an in vitro active composite layer.Proc Natl Acad Sci U S A. 2016 Mar 22;113(12):E1645-54. doi: 10.1073/pnas.1514030113. Epub 2016 Feb 29. Proc Natl Acad Sci U S A. 2016. PMID: 26929326 Free PMC article.
-
Electron crystallography for structural and functional studies of membrane proteins.J Electron Microsc (Tokyo). 2011;60 Suppl 1:S149-59. doi: 10.1093/jmicro/dfr033. J Electron Microsc (Tokyo). 2011. PMID: 21844586 Review.
-
Rafts: scale-dependent, active lipid organization at the cell surface.Traffic. 2004 Apr;5(4):231-40. doi: 10.1111/j.1600-0854.2004.00172.x. Traffic. 2004. PMID: 15030564 Review.
-
Metabolically Biotinylated Reporters for Electron Microscopic Imaging of Cytoplasmic Membrane Microdomains.Methods Mol Biol. 2016;1376:87-96. doi: 10.1007/978-1-4939-3170-5_8. Methods Mol Biol. 2016. PMID: 26552677
-
Membrane sorting in the endocytic and phagocytic pathway of Dictyostelium discoideum.Eur J Cell Biol. 2001 Dec;80(12):754-64. doi: 10.1078/0171-9335-00215. Eur J Cell Biol. 2001. PMID: 11831389
Cited by
-
Phosphorylation of nephrin induces phase separated domains that move through actomyosin contraction.Mol Biol Cell. 2019 Nov 15;30(24):2996-3012. doi: 10.1091/mbc.E18-12-0823. Epub 2019 Oct 10. Mol Biol Cell. 2019. PMID: 31599693 Free PMC article.
-
Toward a new picture of the living plasma membrane.Protein Sci. 2020 Jun;29(6):1355-1365. doi: 10.1002/pro.3874. Epub 2020 May 22. Protein Sci. 2020. PMID: 32297381 Free PMC article. Review.
-
The mystery of membrane organization: composition, regulation and roles of lipid rafts.Nat Rev Mol Cell Biol. 2017 Jun;18(6):361-374. doi: 10.1038/nrm.2017.16. Epub 2017 Mar 30. Nat Rev Mol Cell Biol. 2017. PMID: 28356571 Free PMC article. Review.
-
Domain Stability in Biomimetic Membranes Driven by Lipid Polyunsaturation.J Phys Chem B. 2016 Nov 23;120(46):11930-11941. doi: 10.1021/acs.jpcb.6b06815. Epub 2016 Nov 10. J Phys Chem B. 2016. PMID: 27797198 Free PMC article.
-
Artificial neural networks for the inverse design of nanoparticles with preferential nano-bio behaviors.J Chem Phys. 2020 Aug 7;153(5):054102. doi: 10.1063/5.0013990. J Chem Phys. 2020. PMID: 32770917 Free PMC article.
References
-
- Quote originally attributed to former Saudi oil minister. Sheik Ahmed Zaki Yamani ca 1970s.
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

