Programming a Human Commensal Bacterium, Bacteroides thetaiotaomicron, to Sense and Respond to Stimuli in the Murine Gut Microbiota
- PMID: 26918244
- PMCID: PMC4762051
- DOI: 10.1016/j.cels.2015.06.001
Programming a Human Commensal Bacterium, Bacteroides thetaiotaomicron, to Sense and Respond to Stimuli in the Murine Gut Microbiota
Abstract
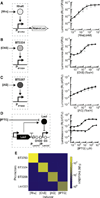
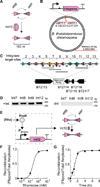
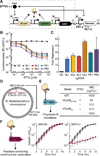
Engineering commensal organisms for challenging applications, such as modulating the gut ecosystem, is hampered by the lack of genetic parts. Here, we describe promoters, ribosome-binding sites, and inducible systems for use in the commensal bacterium Bacteroides thetaiotaomicron, a prevalent and stable resident of the human gut. We achieve up to 10,000-fold range in constitutive gene expression and 100-fold regulation of gene expression with inducible promoters and use these parts to record DNA-encoded memory in the genome. We use CRISPR interference (CRISPRi) for regulated knockdown of recombinant and endogenous gene expression to alter the metabolic capacity of B. thetaiotaomicron and its resistance to antimicrobial peptides. Finally, we show that inducible CRISPRi and recombinase systems can function in B. thetaiotaomicron colonizing the mouse gut. These results provide a blueprint for engineering new chassis and a resource to engineer Bacteroides for surveillance of or therapeutic delivery to the gut microbiome.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

