Chromatin looping as a target for altering erythroid gene expression
- PMID: 26918894
- PMCID: PMC4870130
- DOI: 10.1111/nyas.13012
Chromatin looping as a target for altering erythroid gene expression
Abstract
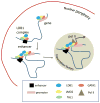
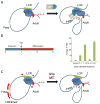
The β-hemoglobinopathies are the most common monogenic disorders in humans, with symptoms arising after birth when the fetal γ-globin genes are silenced and the adult β-globin gene is activated. There is a growing appreciation that genome organization and the folding of chromosomes are key determinants of gene transcription. Underlying this function is the activity of transcriptional enhancers that increase the transcription of target genes over long linear distances. To accomplish this, enhancers engage in close physical contact with target promoters through chromosome folding or looping that is orchestrated by protein complexes that bind to both sites and stabilize their interaction. We find that enhancer activity can be redirected with concomitant changes in gene transcription. Both targeting the β-globin locus control region (LCR) to the γ-globin gene in adult erythroid cells by tethering and epigenetic unmasking of a silenced γ-globin gene lead to increased frequency of LCR/γ-globin contacts and reduced LCR/β-globin contacts. The outcome of these manipulations is robust, pancellular γ-globin transcription activation with a concomitant reduction in β-globin transcription. These examples show that chromosome looping may be considered a therapeutic target for gene activation in β-thalassemia and sickle cell disease.
Keywords: chromatin looping; enhancer; fetal hemoglobin; globin switching; thalassemia.
Published 2016. This article is a U.S. Government work and is in the public domain in the USA.
Conflict of interest statement
The authors declare no conflicts of interest
Figures

References
-
- Bonn S, Zinzen RP, Girardot C, Gustafson EH, Perez-Gonzalez A, Delhomme N, Ghavi-Helm Y, Wilczynski B, Riddell A, Furlong EE. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat Genet. 2012;44(2):148–156. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

