Optimised Pre-Analytical Methods Improve KRAS Mutation Detection in Circulating Tumour DNA (ctDNA) from Patients with Non-Small Cell Lung Cancer (NSCLC)
- PMID: 26918901
- PMCID: PMC4769175
- DOI: 10.1371/journal.pone.0150197
Optimised Pre-Analytical Methods Improve KRAS Mutation Detection in Circulating Tumour DNA (ctDNA) from Patients with Non-Small Cell Lung Cancer (NSCLC)
Abstract
Introduction: Non-invasive mutation testing using circulating tumour DNA (ctDNA) is an attractive premise. This could enable patients without available tumour sample to access more treatment options.
Materials & methods: Peripheral blood and matched tumours were analysed from 45 NSCLC patients. We investigated the impact of pre-analytical variables on DNA yield and/or KRAS mutation detection: sample collection tube type, incubation time, centrifugation steps, plasma input volume and DNA extraction kits.
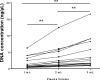
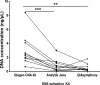
Results: 2 hr incubation time and double plasma centrifugation (2000 x g) reduced overall DNA yield resulting in lowered levels of contaminating genomic DNA (gDNA). Reduced "contamination" and increased KRAS mutation detection was observed using cell-free DNA Blood Collection Tubes (cfDNA BCT) (Streck), after 72 hrs following blood draw compared to EDTA tubes. Plasma input volume and use of different DNA extraction kits impacted DNA yield.
Conclusion: This study demonstrated that successful ctDNA recovery for mutation detection in NSCLC is dependent on pre-analytical steps. Development of standardised methods for the detection of KRAS mutations from ctDNA specimens is recommended to minimise the impact of pre-analytical steps on mutation detection rates. Where rapid sample processing is not possible the use of cfDNA BCT tubes would be advantageous.
Conflict of interest statement
Figures

Similar articles
-
Optimization of circulating cell-free DNA recovery for KRAS mutation and HPV detection in plasma.Cancer Biomark. 2013;13(5):385-94. doi: 10.3233/CBM-130371. Cancer Biomark. 2013. PMID: 24440979
-
Performance of Streck cfDNA Blood Collection Tubes for Liquid Biopsy Testing.PLoS One. 2016 Nov 10;11(11):e0166354. doi: 10.1371/journal.pone.0166354. eCollection 2016. PLoS One. 2016. PMID: 27832189 Free PMC article.
-
Hybrid Capture-Based Genomic Profiling of Circulating Tumor DNA from Patients with Advanced Non-Small Cell Lung Cancer.J Thorac Oncol. 2019 Feb;14(2):255-264. doi: 10.1016/j.jtho.2018.10.008. Epub 2018 Oct 24. J Thorac Oncol. 2019. PMID: 30368012
-
[What future for circulating tumor DNA? Current data and prospects in colorectal, non-small cell lung and pancreatic cancers].Bull Cancer. 2016 Jan;103(1):55-65. doi: 10.1016/j.bulcan.2015.10.017. Epub 2016 Jan 11. Bull Cancer. 2016. PMID: 26790710 Review. French.
-
Cell-free circulating tumor DNA in plasma/serum of non-small cell lung cancer.Tumour Biol. 2015 Jan;36(1):7-19. doi: 10.1007/s13277-014-2758-3. Epub 2014 Oct 29. Tumour Biol. 2015. PMID: 25352029 Review.
Cited by
-
Reproducibility of Digital PCR Assays for Circulating Tumor DNA Analysis in Advanced Breast Cancer.PLoS One. 2016 Oct 19;11(10):e0165023. doi: 10.1371/journal.pone.0165023. eCollection 2016. PLoS One. 2016. PMID: 27760227 Free PMC article.
-
Liquid biopsy in oncology: a consensus statement of the Spanish Society of Pathology and the Spanish Society of Medical Oncology.Clin Transl Oncol. 2020 Jun;22(6):823-834. doi: 10.1007/s12094-019-02211-x. Epub 2019 Sep 26. Clin Transl Oncol. 2020. PMID: 31559582 Free PMC article.
-
Effects of Different Centrifugation Protocols on the Detection of EGFR Mutations in Plasma Cell-Free DNA.Am J Clin Pathol. 2022 Aug 4;158(2):206-211. doi: 10.1093/ajcp/aqac024. Am J Clin Pathol. 2022. PMID: 35285877 Free PMC article.
-
Cell-free DNA in cancer: current insights.Cell Oncol (Dordr). 2019 Feb;42(1):13-28. doi: 10.1007/s13402-018-0413-5. Epub 2018 Oct 26. Cell Oncol (Dordr). 2019. PMID: 30367445 Review.
-
Precision cell-free DNA extraction for liquid biopsy by integrated microfluidics.NPJ Precis Oncol. 2020 Feb 24;4:3. doi: 10.1038/s41698-019-0107-0. eCollection 2020. NPJ Precis Oncol. 2020. PMID: 32133418 Free PMC article.
References
-
- Peake, Mick. National Lung Cancer Audit Report 2014. http://www.hqip.org.uk/. 2014.
-
- Gormally E, Caboux E, Vineis P, Hainaut P. Circulating free DNA in plasma or serum as biomarker of carcinogenesis: Practical aspects and biological significance. 2007;635: 105–117. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

