High-Resolution Analysis of Coronavirus Gene Expression by RNA Sequencing and Ribosome Profiling
- PMID: 26919232
- PMCID: PMC4769073
- DOI: 10.1371/journal.ppat.1005473
High-Resolution Analysis of Coronavirus Gene Expression by RNA Sequencing and Ribosome Profiling
Abstract
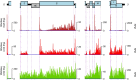
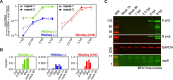
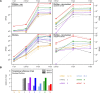
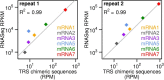
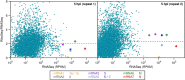
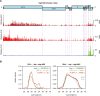
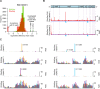
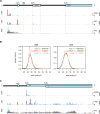
Members of the family Coronaviridae have the largest genomes of all RNA viruses, typically in the region of 30 kilobases. Several coronaviruses, such as Severe acute respiratory syndrome-related coronavirus (SARS-CoV) and Middle East respiratory syndrome-related coronavirus (MERS-CoV), are of medical importance, with high mortality rates and, in the case of SARS-CoV, significant pandemic potential. Other coronaviruses, such as Porcine epidemic diarrhea virus and Avian coronavirus, are important livestock pathogens. Ribosome profiling is a technique which exploits the capacity of the translating ribosome to protect around 30 nucleotides of mRNA from ribonuclease digestion. Ribosome-protected mRNA fragments are purified, subjected to deep sequencing and mapped back to the transcriptome to give a global "snap-shot" of translation. Parallel RNA sequencing allows normalization by transcript abundance. Here we apply ribosome profiling to cells infected with Murine coronavirus, mouse hepatitis virus, strain A59 (MHV-A59), a model coronavirus in the same genus as SARS-CoV and MERS-CoV. The data obtained allowed us to study the kinetics of virus transcription and translation with exquisite precision. We studied the timecourse of positive and negative-sense genomic and subgenomic viral RNA production and the relative translation efficiencies of the different virus ORFs. Virus mRNAs were not found to be translated more efficiently than host mRNAs; rather, virus translation dominates host translation at later time points due to high levels of virus transcripts. Triplet phasing of the profiling data allowed precise determination of translated reading frames and revealed several translated short open reading frames upstream of, or embedded within, known virus protein-coding regions. Ribosome pause sites were identified in the virus replicase polyprotein pp1a ORF and investigated experimentally. Contrary to expectations, ribosomes were not found to pause at the ribosomal frameshift site. To our knowledge this is the first application of ribosome profiling to an RNA virus.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- Corman VM, Baldwin HJ, Fumie Tateno A, Melim Zerbinati R, Annan A, Owusu M, Nkrumah EE, Maganga GD, Oppong S, Adu-Sarkodie Y, Vallo P, da Silva Filho LV, Leroy EM, Thiel V, van der Hoek L, Poon LL, Tschapka M, Drosten C, Drexler JF (2015) Evidence for an ancestral association of human coronavirus 229E with bats. J Virol 89: 11858–11870. 10.1128/JVI.01755-15 - DOI - PMC - PubMed
-
- Bredenbeek PJ, Pachuk CJ, Noten AF, Charité J, Luytjes W, Weiss SR, Spaan WJ (1990) The primary structure and expression of the second open reading frame of the polymerase gene of the coronavirus MHV-A59; a highly conserved polymerase is expressed by an efficient ribosomal frameshifting mechanism. Nucleic Acids Res 18: 1825–32. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

