Molecular Diagnostics of Gliomas Using Next Generation Sequencing of a Glioma-Tailored Gene Panel
- PMID: 26919320
- PMCID: PMC8029406
- DOI: 10.1111/bpa.12367
Molecular Diagnostics of Gliomas Using Next Generation Sequencing of a Glioma-Tailored Gene Panel
Abstract
Current classification of gliomas is based on histological criteria according to the World Health Organization (WHO) classification of tumors of the central nervous system. Over the past years, characteristic genetic profiles have been identified in various glioma types. These can refine tumor diagnostics and provide important prognostic and predictive information. We report on the establishment and validation of gene panel next generation sequencing (NGS) for the molecular diagnostics of gliomas. We designed a glioma-tailored gene panel covering 660 amplicons derived from 20 genes frequently aberrant in different glioma types. Sensitivity and specificity of glioma gene panel NGS for detection of DNA sequence variants and copy number changes were validated by single gene analyses. NGS-based mutation detection was optimized for application on formalin-fixed paraffin-embedded tissue specimens including small stereotactic biopsy samples. NGS data obtained in a retrospective analysis of 121 gliomas allowed for their molecular classification into distinct biological groups, including (i) isocitrate dehydrogenase gene (IDH) 1 or 2 mutant astrocytic gliomas with frequent α-thalassemia/mental retardation syndrome X-linked (ATRX) and tumor protein p53 (TP53) gene mutations, (ii) IDH mutant oligodendroglial tumors with 1p/19q codeletion, telomerase reverse transcriptase (TERT) promoter mutation and frequent Drosophila homolog of capicua (CIC) gene mutation, as well as (iii) IDH wildtype glioblastomas with frequent TERT promoter mutation, phosphatase and tensin homolog (PTEN) mutation and/or epidermal growth factor receptor (EGFR) amplification. Oligoastrocytic gliomas were genetically assigned to either of these groups. Our findings implicate gene panel NGS as a promising diagnostic technique that may facilitate integrated histological and molecular glioma classification.
Keywords: glioma; molecular diagnostics; mutation; next generation sequencing.
© 2016 International Society of Neuropathology.
Figures
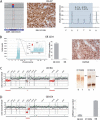
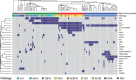

References
-
- Appin CL, Brat DJ (2015) Molecular pathways in gliomagenesis and their relevance to neuropathologic diagnosis. Adv Anat Pathol 22:50–58. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

