New Pseudomonas spp. Are Pathogenic to Citrus
- PMID: 26919540
- PMCID: PMC4769151
- DOI: 10.1371/journal.pone.0148796
New Pseudomonas spp. Are Pathogenic to Citrus
Abstract
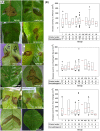
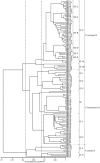
Five putative novel Pseudomonas species shown to be pathogenic to citrus have been characterized in a screening of 126 Pseudomonas strains isolated from diseased citrus leaves and stems in northern Iran. The 126 strains were studied using a polyphasic approach that included phenotypic characterizations and phylogenetic multilocus sequence analysis. The pathogenicity of these strains against 3 cultivars of citrus is demonstrated in greenhouse and field studies. The strains were initially grouped phenotypically and by their partial rpoD gene sequences into 11 coherent groups in the Pseudomonas fluorescens phylogenetic lineage. Fifty-three strains that are representatives of the 11 groups were selected and analyzed by partial sequencing of their 16S rRNA and gyrB genes. The individual and concatenated partial sequences of the three genes were used to construct the corresponding phylogenetic trees. The majority of the strains were identified at the species level: P. lurida (5 strains), P. monteilii (2 strains), P. moraviensis (1 strain), P. orientalis (16 strains), P. simiae (7 strains), P. syringae (46 strains, distributed phylogenetically in at least 5 pathovars), and P. viridiflava (2 strains). This is the first report of pathogenicity on citrus of P. orientalis, P. simiae, P. lurida, P. moraviensis and P. monteilii strains. The remaining 47 strains that could not be identified at the species level are considered representatives of at least 5 putative novel Pseudomonas species that are not yet described.
Conflict of interest statement
Figures



References
-
- Young JM. Taxonomy of Pseudomonas syringae. J Plant Pathol. 2010; 92,S1.5–S1.14
-
- Mulet M, Gomila M, Scotta C, Sánchez D, Lalucat J, García-Valdés E. Concordance between whole-cell matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry and multilocus sequence analysis approaches in species discrimination within the genus Pseudomonas. Syst Appl Microbiol. 2012; 35, 455–464. 10.1016/j.syapm.2012.08.007 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

