An integrative analysis of reprogramming in human isogenic system identified a clone selection criterion
- PMID: 26919644
- PMCID: PMC4889246
- DOI: 10.1080/15384101.2016.1152425
An integrative analysis of reprogramming in human isogenic system identified a clone selection criterion
Abstract
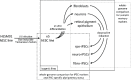
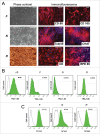
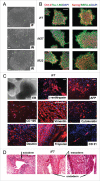
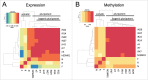
The pluripotency of newly developed human induced pluripotent stem cells (iPSCs) is usually characterized by physiological parameters; i.e., by their ability to maintain the undifferentiated state and to differentiate into derivatives of the 3 germ layers. Nevertheless, a molecular comparison of physiologically normal iPSCs to the "gold standard" of pluripotency, embryonic stem cells (ESCs), often reveals a set of genes with different expression and/or methylation patterns in iPSCs and ESCs. To evaluate the contribution of the reprogramming process, parental cell type, and fortuity in the signature of human iPSCs, we developed a complete isogenic reprogramming system. We performed a genome-wide comparison of the transcriptome and the methylome of human isogenic ESCs, 3 types of ESC-derived somatic cells (fibroblasts, retinal pigment epithelium and neural cells), and 3 pairs of iPSC lines derived from these somatic cells. Our analysis revealed a high input of stochasticity in the iPSC signature that does not retain specific traces of the parental cell type and reprogramming process. We showed that 5 iPSC clones are sufficient to find with 95% confidence at least one iPSC clone indistinguishable from their hypothetical isogenic ESC line. Additionally, on the basis of a small set of genes that are characteristic of all iPSC lines and isogenic ESCs, we formulated an approach of "the best iPSC line" selection and confirmed it on an independent dataset.
Keywords: DNA methylation; gene transcription; genome-wide analysis; human pluripotent stem cells; isogenic; reprogramming; somatic memory.
Figures

References
-
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science 1998; 282:1145-7; PMID:9804556; http://dx.doi.org/10.1126/science.282.5391.1145 - DOI - PubMed
-
- Pan CK, Heilweil G, Lanza R, Schwartz SD. Embryonic stem cells as a treatment for macular degeneration. Expert Opin Biol Ther 2013; 13:1125-33; PMID:23705996; http://dx.doi.org/10.1517/14712598.2013.793304 - DOI - PubMed
-
- Schwartz SD, Hubschman J-P, Heilwell G, Franco-Cardenas V, Pan CK, Ostrick RM, Mickunas E, Gay R, Klimanskaya I, Lanza R. Embryonic stem cell trials for macular degeneration: a preliminary report. Lancet 2012; 379:713-20; PMID:22281388; http://dx.doi.org/10.1016/S0140-6736(12)60028-2 - DOI - PubMed
-
- Bock C, Kiskinis E, Verstappen G, Gu H, Boulting G, Smith ZD, Ziller M, Croft GF, Amoroso MW, Oakley DH, et al.. Reference Maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell 2011; 144:439-52; PMID:21295703; http://dx.doi.org/10.1016/j.cell.2010.12.032 - DOI - PMC - PubMed
-
- Marchetto MCN, Yeo GW, Kainohana O, Marsala M, Gage FH, Muotri AR. Transcriptional signature and memory retention of human-induced pluripotent stem cells. PLoS One 2009; 4:e7076; PMID:19763270; http://dx.doi.org/10.1371/journal.pone.0007076 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
