Changes in Proteome Profile of Peripheral Blood Mononuclear Cells in Chronic Chagas Disease
- PMID: 26919708
- PMCID: PMC4769231
- DOI: 10.1371/journal.pntd.0004490
Changes in Proteome Profile of Peripheral Blood Mononuclear Cells in Chronic Chagas Disease
Abstract
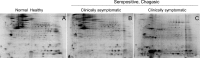
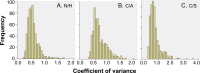
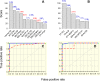
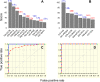
Trypanosoma cruzi (Tc) infection causes chagasic cardiomyopathy; however, why 30-40% of the patients develop clinical disease is not known. To discover the pathomechanisms in disease progression, we obtained the proteome signature of peripheral blood mononuclear cells (PBMCs) of normal healthy controls (N/H, n = 30) and subjects that were seropositive for Tc-specific antibodies, but were clinically asymptomatic (C/A, n = 25) or clinically symptomatic (C/S, n = 28) with cardiac involvement and left ventricular dysfunction. Protein samples were labeled with BODIPY FL-maleimide (dynamic range: > 4 orders of magnitude, detection limit: 5 f-mol) and resolved by two-dimensional gel electrophoresis (2D-GE). After normalizing the gel images, protein spots that exhibited differential abundance in any of the two groups were analyzed by mass spectrometry, and searched against UniProt human database for protein identification. We found 213 and 199 protein spots (fold change: |≥ 1.5|, p< 0.05) were differentially abundant in C/A and C/S individuals, respectively, with respect to N/H controls. Ingenuity Pathway Analysis (IPA) of PBMCs proteome dataset identified an increase in disorganization of cytoskeletal assembly and recruitment/activation and migration of immune cells in all chagasic subjects, though the invasion capacity of cells was decreased in C/S individuals. IPA predicted with high probability a decline in cell survival and free radical scavenging capacity in C/S (but not C/A) subjects. The MYC/SP1 transcription factors that regulate hypoxia and oxidative/inflammatory stress were predicted to be key targets in the context of control of Chagas disease severity. Further, MARS-modeling identified a panel of proteins that had >93% prediction success in classifying infected individuals with no disease and those with cardiac involvement and LV dysfunction. In conclusion, we have identified molecular pathways and a panel of proteins that could aid in detecting seropositive individuals at risk of developing cardiomyopathy.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- World Health Organization (2010) Chagas disease: control and elimination UNDP/World Bank/WHO, http://apps.who.int/gb/ebwha/pdf_files/WHA63/A63_17-en.pdf, accessed on Jan 15, 2016.
-
- de Assis GF, Azeredo BV, Carbajal de la Fuente AL, Diotaiuti L, de Lana M (2007) Domiciliation of Triatoma pseudomaculata (Correa e Espinola 1964) in the Jequitinhonha Valley, State of Minas Gerais. Rev Soc Bras Med Trop 40: 391–396. - PubMed
-
- Garraud O, Andreu G, Elghouzzi MH, Laperche S, Lefrere JJ (2007) Measures to prevent transfusion-associated protozoal infections in non-endemic countries. Travel Med Infect Dis 5: 110–112. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

