Naturally Occurring Isoleucyl-tRNA Synthetase without tRNA-dependent Pre-transfer Editing
- PMID: 26921320
- PMCID: PMC4861432
- DOI: 10.1074/jbc.M115.698225
Naturally Occurring Isoleucyl-tRNA Synthetase without tRNA-dependent Pre-transfer Editing
Abstract
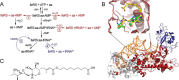
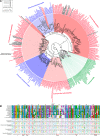
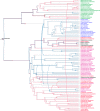
Isoleucyl-tRNA synthetase (IleRS) is unusual among aminoacyl-tRNA synthetases in having a tRNA-dependent pre-transfer editing activity. Alongside the typical bacterial IleRS (such as Escherichia coli IleRS), some bacteria also have the enzymes (eukaryote-like) that cluster with eukaryotic IleRSs and exhibit low sensitivity to the antibiotic mupirocin. Our phylogenetic analysis suggests that the ileS1 and ileS2 genes of contemporary bacteria are the descendants of genes that might have arisen by an ancient duplication event before the separation of bacteria and archaea. We present the analysis of evolutionary constraints of the synthetic and editing reactions in eukaryotic/eukaryote-like IleRSs, which share a common origin but diverged through adaptation to different cell environments. The enzyme from the yeast cytosol exhibits tRNA-dependent pre-transfer editing analogous to E. coli IleRS. This argues for the presence of this proofreading in the common ancestor of both IleRS types and an ancient origin of the synthetic site-based quality control step. Yet surprisingly, the eukaryote-like enzyme from Streptomyces griseus IleRS lacks this capacity; at the same time, its synthetic site displays the 10(3)-fold drop in sensitivity to antibiotic mupirocin relative to the yeast enzyme. The discovery that pre-transfer editing is optional in IleRSs lends support to the notion that the conserved post-transfer editing domain is the main checkpoint in these enzymes. We substantiated this by showing that under error-prone conditions S. griseus IleRS is able to rescue the growth of an E. coli lacking functional IleRS, providing the first evidence that tRNA-dependent pre-transfer editing in IleRS is not essential for cell viability.
Keywords: aminoacyl tRNA synthetase; antibiotic resistance; isoleucyl-tRNA synthetase; mupirocin; proofreading; protein evolution; protein synthesis; tRNA-dependent pre-transfer editing; transfer RNA (tRNA).
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures
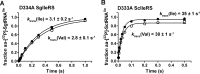


References
-
- Perona J. J., and Gruic-Sovulj I. (2014) Synthetic and editing mechanisms of aminoacyl-tRNA synthetases. Top. Curr. Chem. 344, 1–41 - PubMed
-
- Jakubowski H. (2012) Quality control in tRNA charging. Wiley Interdiscip. Rev. RNA 3, 295–310 - PubMed
-
- Nordin B. E., and Schimmel P. (2005) in Aminoacyl tRNA Synthetases (Ibba M., Francklyn C., and Cusack S., eds) pp. 24–35, Landes Biosciences, Georgetown, TX
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

