Autophagy in acute kidney injury
- PMID: 26924060
- PMCID: PMC4801755
- DOI: 10.1016/j.kint.2015.11.021
Autophagy in acute kidney injury
Abstract
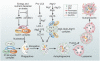
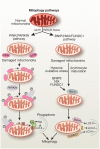
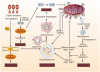
Autophagy is a conserved multistep pathway that degrades and recycles damaged organelles and macromolecules to maintain intracellular homeostasis. The autophagy pathway is upregulated under stress conditions including cell starvation, hypoxia, nutrient and growth-factor deprivation, endoplasmic reticulum stress, and oxidant injury, most of which are involved in the pathogenesis of acute kidney injury (AKI). Recent studies demonstrate that basal autophagy in the kidney is vital for the normal homeostasis of the proximal tubules. Deletion of key autophagy proteins impaired renal function and increased p62 levels and oxidative stress. In models of AKI, autophagy deletion in proximal tubules worsened tubular injury and renal function, highlighting that autophagy is renoprotective in models of AKI. In addition to nonselective sequestration of autophagic cargo, autophagy can facilitate selective degradation of damaged organelles, particularly mitochondrial degradation through the process of mitophagy. Damaged mitochondria accumulate in autophagy-deficient kidneys of mice subjected to ischemia-reperfusion injury, but the precise mechanisms of regulation of mitophagy in AKI are not yet elucidated. Recent progress in identifying the interplay of autophagy, apoptosis, and regulated necrosis has revived interest in examining shared pathways/molecules in this crosstalk during the pathogenesis of AKI. Autophagy and its associated pathways pose potentially unique targets for therapeutic interventions in AKI.
Keywords: acute kidney injury; apoptosis; autophagy; cell death; cisplatin nephrotoxicity; ischemia reperfusion; mitophagy.
Published by Elsevier Inc.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

