The Coding Region of the HCV Genome Contains a Network of Regulatory RNA Structures
- PMID: 26924328
- PMCID: PMC4826301
- DOI: 10.1016/j.molcel.2016.01.024
The Coding Region of the HCV Genome Contains a Network of Regulatory RNA Structures
Abstract
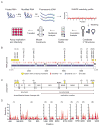
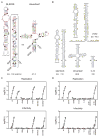
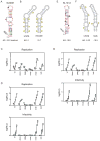
RNA is a versatile macromolecule that accommodates functional information in primary sequence and secondary and tertiary structure. We use a combination of chemical probing, RNA structure modeling, comparative sequence analysis, and functional assays to examine the role of RNA structure in the hepatitis C virus (HCV) genome. We describe a set of conserved but functionally diverse structural RNA motifs that occur in multiple coding regions of the HCV genome, and we demonstrate that conformational changes in these motifs influence specific stages in the virus' life cycle. Our study shows that these types of structures can pervade a genome, where they play specific mechanistic and regulatory roles, constituting a "code within the code" for controlling biological processes.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures

References
-
- Bartenschlager R, Frese M, Pietschmann T. Novel insights into hepatitis C virus replication and persistence. Adv Virus Res. 2004;63:71–180. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

