Down Syndrome Developmental Brain Transcriptome Reveals Defective Oligodendrocyte Differentiation and Myelination
- PMID: 26924435
- PMCID: PMC4795969
- DOI: 10.1016/j.neuron.2016.01.042
Down Syndrome Developmental Brain Transcriptome Reveals Defective Oligodendrocyte Differentiation and Myelination
Abstract
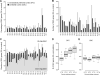
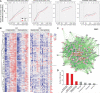
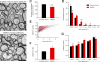
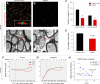
Trisomy 21, or Down syndrome (DS), is the most common genetic cause of developmental delay and intellectual disability. To gain insight into the underlying molecular and cellular pathogenesis, we conducted a multi-region transcriptome analysis of DS and euploid control brains spanning from mid-fetal development to adulthood. We found genome-wide alterations in the expression of a large number of genes, many of which exhibited temporal and spatial specificity and were associated with distinct biological processes. In particular, we uncovered co-dysregulation of genes associated with oligodendrocyte differentiation and myelination that were validated via cross-species comparison to Ts65Dn trisomy mice. Furthermore, we show that hypomyelination present in Ts65Dn mice is in part due to cell-autonomous effects of trisomy on oligodendrocyte differentiation and results in slower neocortical action potential transmission. Together, these results identify defects in white matter development and function in DS, and they provide a transcriptional framework for further investigating DS neuropathogenesis.
Keywords: brain development; gene expression; genomics; glia; neocortex; neurodevelopmental disorders; white matter.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures




References
-
- Abraham H, Vincze A, Veszpremi B, Kravjak A, Gomori E, Kovacs GG, Seress L. Impaired myelination of the human hippocampal formation in Down syndrome. Int J Dev Neurosci. 2012;30:147–158. - PubMed
-
- Bahn S, Mimmack M, Ryan M, Caldwell MA, Jauniaux E, Starkey M, Svendsen CN, Emson P. Neuronal target genes of the neuron-restrictive silencer factor in neurospheres derived from fetuses with Down's syndrome: a gene expression study. Lancet. 2002;359:310–315. - PubMed
-
- Bartzokis G. Acetylcholinesterase inhibitors may improve myelin integrity. Biol. Psychiatry. 2007;62:294–301. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
