Mechanisms underlying structural variant formation in genomic disorders
- PMID: 26924765
- PMCID: PMC4827625
- DOI: 10.1038/nrg.2015.25
Mechanisms underlying structural variant formation in genomic disorders
Abstract
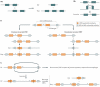
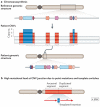
With the recent burst of technological developments in genomics, and the clinical implementation of genome-wide assays, our understanding of the molecular basis of genomic disorders, specifically the contribution of structural variation to disease burden, is evolving quickly. Ongoing studies have revealed a ubiquitous role for genome architecture in the formation of structural variants at a given locus, both in DNA recombination-based processes and in replication-based processes. These reports showcase the influence of repeat sequences on genomic stability and structural variant complexity and also highlight the tremendous plasticity and dynamic nature of our genome in evolution, health and disease susceptibility.
Figures

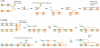

References
-
- Lupski JR. Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 1998;14:417–422. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

