Stimuli Reduce the Dimensionality of Cortical Activity
- PMID: 26924968
- PMCID: PMC4756130
- DOI: 10.3389/fnsys.2016.00011
Stimuli Reduce the Dimensionality of Cortical Activity
Abstract
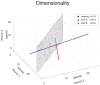
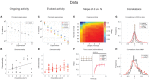
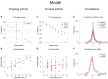
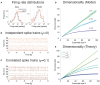
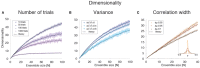
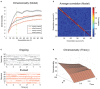
The activity of ensembles of simultaneously recorded neurons can be represented as a set of points in the space of firing rates. Even though the dimension of this space is equal to the ensemble size, neural activity can be effectively localized on smaller subspaces. The dimensionality of the neural space is an important determinant of the computational tasks supported by the neural activity. Here, we investigate the dimensionality of neural ensembles from the sensory cortex of alert rats during periods of ongoing (inter-trial) and stimulus-evoked activity. We find that dimensionality grows linearly with ensemble size, and grows significantly faster during ongoing activity compared to evoked activity. We explain these results using a spiking network model based on a clustered architecture. The model captures the difference in growth rate between ongoing and evoked activity and predicts a characteristic scaling with ensemble size that could be tested in high-density multi-electrode recordings. Moreover, we present a simple theory that predicts the existence of an upper bound on dimensionality. This upper bound is inversely proportional to the amount of pair-wise correlations and, compared to a homogeneous network without clusters, it is larger by a factor equal to the number of clusters. The empirical estimation of such bounds depends on the number and duration of trials and is well predicted by the theory. Together, these results provide a framework to analyze neural dimensionality in alert animals, its behavior under stimulus presentation, and its theoretical dependence on ensemble size, number of clusters, and correlations in spiking network models.
Keywords: dimensionality; gustatory cortex; hidden markov models; mean field theory; metastable dynamics; ongoing activity; spiking network model.
Figures



References
-
- Abbott L. F., Rajan K., Sompolinsky H. (2011). Interactions between intrinsic and stimulus-evoked activity in recurrent neural networks, in The Dynamic Brain: An Exploration of Neuronal Variability and its Functional Significance, eds Glanzman D. L., Ding M. (New York, NY: Oxford University Press; ), 65–82.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

