Two LcbHLH Transcription Factors Interacting with LcMYB1 in Regulating Late Structural Genes of Anthocyanin Biosynthesis in Nicotiana and Litchi chinensis During Anthocyanin Accumulation
- PMID: 26925082
- PMCID: PMC4757707
- DOI: 10.3389/fpls.2016.00166
Two LcbHLH Transcription Factors Interacting with LcMYB1 in Regulating Late Structural Genes of Anthocyanin Biosynthesis in Nicotiana and Litchi chinensis During Anthocyanin Accumulation
Abstract
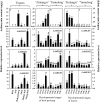
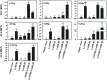
Anthocyanin biosynthesis requires the MYB-bHLH-WD40 protein complex to activate the late biosynthetic genes. LcMYB1 was thought to act as key regulator in anthocyanin biosynthesis of litchi. However, basic helix-loop-helix proteins (bHLHs) as partners have not been identified yet. The present study describes the functional characterization of three litchi bHLH candidate anthocyanin regulators, LcbHLH1, LcbHLH2, and LcbHLH3. Although these three litchi bHLHs phylogenetically clustered with bHLH proteins involved in anthcoyanin biosynthesis in other plant, only LcbHLH1 and LcbHLH3 were found to localize in the nucleus and physically interact with LcMYB1. The transcription levels of all these bHLHs were not coordinated with anthocyanin accumulation in different tissues and during development. However, when co-infiltrated with LcMYB1, both LcbHLH1 and LcbHLH3 enhanced anthocyanin accumulation in tobacco leaves with LcbHLH3 being the best inducer. Significant accumulation of anthocyanins in leaves transformed with the combination of LcMYB1 and LcbHLH3 were noticed, and this was associated with the up-regulation of two tobacco endogenous bHLH regulators, NtAn1a and NtAn1b, and late structural genes, like NtDFR and NtANS. Significant activity of the ANS promoter was observed in transient expression assays either with LcMYB1-LcbHLH1 or LcMYB1-LcbHLH3, while only minute activity was detected after transformation with only LcMYB1. In contrast, no activity was measured after induction with the combination of LcbHLH2 and LcMYB1. Higher DFR expression was also oberseved in paralleling with higher anthocyanins in co-transformed lines. LcbHLH1 and LcbHLH3 are essential partner of LcMYB1 in regulating the anthocyanin production in tobacco and probably also in litchi. The LcMYB1-LcbHLH complex enhanced anthocyanin accumulation may associate with activating the transcription of DFR and ANS.
Keywords: Litchi chinensis; MYB; anthocyanins; bHLH; interaction; tobacco.
Figures







References
LinkOut - more resources
Full Text Sources
Other Literature Sources

