RNA-Seq of the Caribbean reef-building coral Orbicella faveolata (Scleractinia-Merulinidae) under bleaching and disease stress expands models of coral innate immunity
- PMID: 26925311
- PMCID: PMC4768675
- DOI: 10.7717/peerj.1616
RNA-Seq of the Caribbean reef-building coral Orbicella faveolata (Scleractinia-Merulinidae) under bleaching and disease stress expands models of coral innate immunity
Abstract
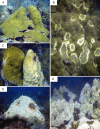
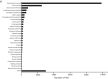
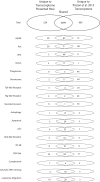
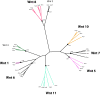
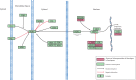
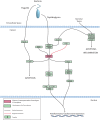
Climate change-driven coral disease outbreaks have led to widespread declines in coral populations. Early work on coral genomics established that corals have a complex innate immune system, and whole-transcriptome gene expression studies have revealed mechanisms by which the coral immune system responds to stress and disease. The present investigation expands bioinformatic data available to study coral molecular physiology through the assembly and annotation of a reference transcriptome of the Caribbean reef-building coral, Orbicella faveolata. Samples were collected during a warm water thermal anomaly, coral bleaching event and Caribbean yellow band disease outbreak in 2010 in Puerto Rico. Multiplex sequencing of RNA on the Illumina GAIIx platform and de novo transcriptome assembly by Trinity produced 70,745,177 raw short-sequence reads and 32,463 O. faveolata transcripts, respectively. The reference transcriptome was annotated with gene ontologies, mapped to KEGG pathways, and a predicted proteome of 20,488 sequences was generated. Protein families and signaling pathways that are essential in the regulation of innate immunity across Phyla were investigated in-depth. Results were used to develop models of evolutionarily conserved Wnt, Notch, Rig-like receptor, Nod-like receptor, and Dicer signaling. O. faveolata is a coral species that has been studied widely under climate-driven stress and disease, and the present investigation provides new data on the genes that putatively regulate its immune system.
Keywords: Cnidaria; Coral; Disease; Innate immunity; RNA-seq.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Ainsworth T, Kvennefors E, Blackall L, Fine M, Hoegh-Guldberg O. Disease and cell death in white syndrome of Acroporid corals on the Great Barrier Reef. Marine Biology. 2007;151(1):19–29. doi: 10.1007/s00227-006-0449-3. - DOI
-
- Anderson D, Gilchrist S. Development of a novel method for coral RNA isolation and the expression of a programmed cell death gene in white plague-diseased Diploria strigosa (Dana, 1846). Proceedings of the 11th International Coral Reef Symposium; Ft. Lauderdale, Florida. 2008. pp. 211–215.
-
- Antonius A. Tenth Meeting of the Association of Island Marine Laboratories of the Caribbean. Mayaguez: University of Puerto Rico; 1973. New observations on coral destruction in reefs [Abstract] p. 3.
LinkOut - more resources
Full Text Sources
Other Literature Sources

