Synergistic action of master transcription factors controls epithelial-to-mesenchymal transition
- PMID: 26926107
- PMCID: PMC4824118
- DOI: 10.1093/nar/gkw126
Synergistic action of master transcription factors controls epithelial-to-mesenchymal transition
Abstract
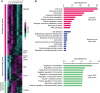
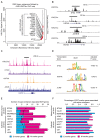
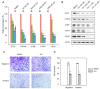
Epithelial-to-mesenchymal transition (EMT) is a complex multistep process in which phenotype switches are mediated by a network of transcription factors (TFs). Systematic characterization of all dynamic TFs controlling EMT state transitions, especially for the intermediate partial-EMT state, represents a highly relevant yet largely unexplored task. Here, we performed a computational analysis that integrated time-course EMT transcriptomic data with public cistromic data and identified three synergistic master TFs (ETS2, HNF4A and JUNB) that regulate the transition through the partial-EMT state. Overexpression of these regulators predicted a poor clinical outcome, and their elimination readily abolished TGF-β-induced EMT. Importantly, these factors utilized a clique motif, physically interact and their cumulative binding generally characterized EMT-associated genes. Furthermore, analyses of H3K27ac ChIP-seq data revealed that ETS2, HNF4A and JUNB are associated with super-enhancers and the administration of BRD4 inhibitor readily abolished TGF-β-induced EMT. These findings have implications for systematic discovery of master EMT regulators and super-enhancers as novel targets for controlling metastasis.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




References
-
- De Craene B., Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat. Rev. Cancer. 2013;13:97–110. - PubMed
-
- Thiery J.P., Acloque H., Huang R.Y., Nieto M.A. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. - PubMed
-
- Yang J., Weinberg R.A. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev. Cell. 2008;14:818–829. - PubMed
-
- Garber K. Epithelial-to-mesenchymal transition is important to metastasis, but questions remain. J. Natl. Cancer Inst. 2008;100:232–233. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

