Critical Genomic Networks and Vasoreactive Variants in Idiopathic Pulmonary Arterial Hypertension
- PMID: 26926454
- PMCID: PMC5003329
- DOI: 10.1164/rccm.201508-1678OC
Critical Genomic Networks and Vasoreactive Variants in Idiopathic Pulmonary Arterial Hypertension
Abstract
Rationale: Idiopathic pulmonary arterial hypertension (IPAH) is usually without an identified genetic cause, despite clinical and molecular similarity to bone morphogenetic protein receptor type 2 mutation-associated heritable pulmonary arterial hypertension (PAH). There is phenotypic heterogeneity in IPAH, with a minority of patients showing long-term improvement with calcium channel-blocker therapy.
Objectives: We sought to identify gene variants (GVs) underlying IPAH and determine whether GVs differ in vasodilator-responsive IPAH (VR-PAH) versus vasodilator-nonresponsive IPAH (VN-PAH).
Methods: We performed whole-exome sequencing (WES) on 36 patients with IPAH: 17 with VR-PAH and 19 with VN-PAH. Wnt pathway differences were explored in human lung fibroblasts.
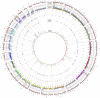
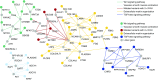
Measurements and main results: We identified 1,369 genes with 1,580 variants unique to IPAH. We used a gene ontology approach to analyze variants and identified overrepresentation of several pathways, including cytoskeletal function and ion binding. By mapping WES data to prior genome-wide association study data, Wnt pathway genes were highlighted. Using the connectivity map to define genetic differences between VR-PAH and VN-PAH, we found enrichment in vascular smooth muscle cell contraction pathways and greater genetic variation in VR-PAH versus VN-PAH. Using human lung fibroblasts, we found increased stimulated Wnt activity in IPAH versus controls.
Conclusions: A pathway-based analysis of WES data in IPAH demonstrated multiple rare GVs that converge on key biological pathways, such as cytoskeletal function and Wnt signaling pathway. Vascular smooth muscle contraction-related genes were enriched in VR-PAH, suggesting a potentially different genetic predisposition for VR-PAH. This pathway-based approach may be applied to next-generation sequencing data in other diseases to uncover the contribution of unexpected or multiple GVs to a phenotype.
Keywords: pulmonary arterial hypertension; vasodilator responsive; whole-exome sequencing.
Figures




Comment in
-
Genetic Insights into Pulmonary Arterial Hypertension. Application of Whole-Exome Sequencing to the Study of Pathogenic Mechanisms.Am J Respir Crit Care Med. 2016 Aug 15;194(4):393-7. doi: 10.1164/rccm.201603-0577ED. Am J Respir Crit Care Med. 2016. PMID: 27525458 Free PMC article. No abstract available.
References
-
- Hoeper MM, Bogaard HJ, Condliffe R, Frantz R, Khanna D, Kurzyna M, Langleben D, Manes A, Satoh T, Torres F, et al. Definitions and diagnosis of pulmonary hypertension. J Am Coll Cardiol. 2013;62:D42–D50. - PubMed
-
- Simonneau G, Gatzoulis MA, Adatia I, Celermajer D, Denton C, Ghofrani A, Gomez Sanchez MA, Krishna Kumar R, Landzberg M, Machado RF, et al. Updated clinical classification of pulmonary hypertension. J Am Coll Cardiol. 2013;62:D34–D41. - PubMed
-
- Galiè N, Corris PA, Frost A, Girgis RE, Granton J, Jing ZC, Klepetko W, McGoon MD, McLaughlin VV, Preston IR, et al. Updated treatment algorithm of pulmonary arterial hypertension. J Am Coll Cardiol. 2013;62:D60–D72. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

