Biochemical Characterization and Complete Conversion of Coenzyme Specificity of Isocitrate Dehydrogenase from Bifidobacterium longum
- PMID: 26927087
- PMCID: PMC4813160
- DOI: 10.3390/ijms17030296
Biochemical Characterization and Complete Conversion of Coenzyme Specificity of Isocitrate Dehydrogenase from Bifidobacterium longum
Abstract
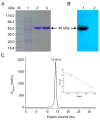
Bifidobacterium longum is a very important gram-positive non-pathogenic bacterium in the human gastrointestinal tract for keeping the digestive and immune system healthy. Isocitrate dehydrogenase (IDH) from B. longum (BlIDH), a novel member in Type II subfamily, was overexpressed, purified and biochemically characterized in detail. The active form of BlIDH was an 83-kDa homodimer. Kinetic analysis showed BlIDH was a NADP⁺-dependent IDH (NADP-IDH), with a 567- and 193-fold preference for NADP⁺ over NAD⁺ in the presence of Mg(2+) and Mn(2+), respectively. The maximal activity for BlIDH occurred at 60 °C (with Mn(2+)) and 65 °C (with Mg(2+)), and pH 7.5 (with Mn(2+)) and pH 8.0 (with Mg(2+)). Heat-inactivation profiles revealed that BlIDH retained 50% of maximal activity after incubation at 45 °C for 20 min with either Mn(2+) or Mg(2+). Furthermore, the coenzyme specificity of BlIDH can be completely reversed from NADP⁺ to NAD⁺ by a factor of 2387 by replacing six residues. This current work, the first report on the coenzyme specificity conversion of Type II NADP-IDHs, would provide better insight into the evolution of NADP⁺ use by the IDH family.
Keywords: Bifidobacterium longum; biochemical characterization; coenzyme specificity determinants; isocitrate dehydrogenase; kinetics.
Figures




References
-
- Miyazaki J., Kobashi N., Nishiyama M., Yamane H. Characterization of homoisocitrate dehydrogenase involved in lysine biosynthesis of an extremely thermophilic bacterium, Thermus thermophilus HB27, and evolutionary implication of β-decarboxylating dehydrogenase. J. Biol. Chem. 2003;278:1864–1871. doi: 10.1074/jbc.M205133200. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

