Disabled homolog 2 controls macrophage phenotypic polarization and adipose tissue inflammation
- PMID: 26927671
- PMCID: PMC4811113
- DOI: 10.1172/JCI79590
Disabled homolog 2 controls macrophage phenotypic polarization and adipose tissue inflammation
Abstract
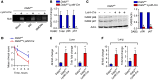
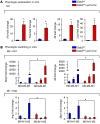
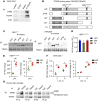
Acute and chronic tissue injury results in the generation of a myriad of environmental cues that macrophages respond to by changing their phenotype and function. This phenotypic regulation is critical for controlling tissue inflammation and resolution. Here, we have identified the adaptor protein disabled homolog 2 (DAB2) as a regulator of phenotypic switching in macrophages. Dab2 expression was upregulated in M2 macrophages and suppressed in M1 macrophages isolated from both mice and humans, and genetic deletion of Dab2 predisposed macrophages to adopt a proinflammatory M1 phenotype. In mice with myeloid cell-specific deletion of Dab2 (Dab2fl/fl Lysm-Cre), treatment with sublethal doses of LPS resulted in increased proinflammatory gene expression and macrophage activation. Moreover, chronic high-fat feeding exacerbated adipose tissue inflammation, M1 polarization of adipose tissue macrophages, and the development of insulin resistance in DAB2-deficient animals compared with controls. Mutational analyses revealed that DAB2 interacts with TNF receptor-associated factor 6 (TRAF6) and attenuates IκB kinase β-dependent (IKKβ-dependent) phosphorylation of Ser536 in the transactivation domain of NF-κB p65. Together, these findings reveal that DAB2 is critical for controlling inflammatory signaling during phenotypic polarization of macrophages and suggest that manipulation of DAB2 expression and function may hold therapeutic potential for the treatment of acute and chronic inflammatory disorders.
Figures




References
-
- Mantovani A, Biswas SK, Galdiero MR, Sica A, Locati M. Macrophage plasticity and polarization in tissue repair and remodelling. J Pathol. 2012;229(2):176–185. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 GM007267/GM/NIGMS NIH HHS/United States
- 5T32AI007496/AI/NIAID NIH HHS/United States
- R01 DK096076/DK/NIDDK NIH HHS/United States
- T32 AI007496/AI/NIAID NIH HHS/United States
- R01DK096076/DK/NIDDK NIH HHS/United States
- R01 DK101946/DK/NIDDK NIH HHS/United States
- HL-33391/HL/NHLBI NIH HHS/United States
- R01 AI015608/AI/NIAID NIH HHS/United States
- R01AI-15608/AI/NIAID NIH HHS/United States
- T32 GM007055/GM/NIGMS NIH HHS/United States
- T32 HL007284/HL/NHLBI NIH HHS/United States
- 5T32 HL007284/HL/NHLBI NIH HHS/United States
- R01 HL084422/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

