DNA methylation levels at individual age-associated CpG sites can be indicative for life expectancy
- PMID: 26928272
- PMCID: PMC4789590
- DOI: 10.18632/aging.100908
DNA methylation levels at individual age-associated CpG sites can be indicative for life expectancy
Abstract
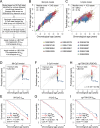
DNA-methylation (DNAm) levels at age-associated CpG sites can be combined into epigenetic aging signatures to estimate donor age. It has been demonstrated that the difference between such epigenetic age-predictions and chronological age is indicative for of all-cause mortality in later life. In this study, we tested alternative epigenetic signatures and followed the hypothesis that even individual age-associated CpG sites might be indicative for life-expectancy. Using a 99-CpG aging model, a five-year higher age-prediction was associated with 11% greater mortality risk in DNAm profiles of the Lothian Birth Cohort 1921 study. However, models based on three CpGs, or even individual CpGs, generally revealed very high offsets in age-predictions if applied to independent microarray datasets. On the other hand, we demonstrate that DNAm levels at several individual age-associated CpGs seem to be associated with life expectancy - e.g., at CpGs associated with the genesPDE4C and CLCN6. Our results support the notion that small aging signatures should rather be analysed by more quantitative methods, such as site-specific pyrosequencing, as the precision of age-predictions is rather low on independent microarray datasets. Nevertheless, the results hold the perspective that simple epigenetic biomarkers, based on few or individual age-associated CpGs, could assist the estimation of biological age.
Keywords: CLCN6; DNA-methylation; PDE4C; age; aging; epigenetic; mortality; prediction; predictor; survival.
Conflict of interest statement
RWTH Aachen University Medical School is has a patent pending for the 3-CpG model for epigenetic age-predictions. W.W. is one of the founders of Cygenia GmbH that may provide service for this method (
Figures

References
-
- Teschendorff AE, Menon U, Gentry-Maharaj A, Ramus SJ, Weisenberger DJ, Shen H, Campan M, Noushmehr H, Bell CG, Maxwell AP, Savage DA, Mueller-Holzner E, Marth C, et al. Age-dependent DNA methylation of genes that are suppressed in stem cells is a hallmark of cancer. Genome Res. 2010;20:440–446. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

