Peroxidase activation of cytoglobin by anionic phospholipids: Mechanisms and consequences
- PMID: 26928591
- PMCID: PMC4821708
- DOI: 10.1016/j.bbalip.2016.02.022
Peroxidase activation of cytoglobin by anionic phospholipids: Mechanisms and consequences
Abstract
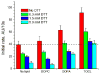
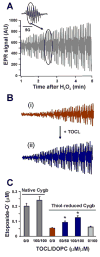
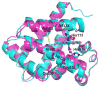
Cytoglobin (Cygb) is a hexa-coordinated hemoprotein with yet to be defined physiological functions. The iron coordination and spin state of the Cygb heme group are sensitive to oxidation of two cysteine residues (Cys38/Cys83) and/or the binding of free fatty acids. However, the roles of redox vs lipid regulators of Cygb's structural rearrangements in the context of the protein peroxidase competence are not known. Searching for physiologically relevant lipid regulators of Cygb, here we report that anionic phospholipids, particularly phosphatidylinositolphosphates, affect structural organization of the protein and modulate its iron state and peroxidase activity both conjointly and/or independently of cysteine oxidation. Thus, different anionic lipids can operate in cysteine-dependent and cysteine-independent ways as inducers of the peroxidase activity. We establish that Cygb's peroxidase activity can be utilized for the catalysis of peroxidation of anionic phospholipids (including phosphatidylinositolphosphates) yielding mono-oxygenated molecular species. Combined with the computational simulations we propose a bipartite lipid binding model that rationalizes the modes of interactions with phospholipids, the effects on structural re-arrangements and the peroxidase activity of the hemoprotein.
Keywords: Cytoglobin; Lipid binding; Peroxidase activity; Phosphatidylinositolphosphates.
Copyright © 2016 Elsevier B.V. All rights reserved.
Figures



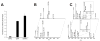
References
-
- Reeder BJ. The redox activity of hemoglobins: from physiologic functions to pathologic mechanisms. Antioxid Redox Signal. 2010;13:1087–123. - PubMed
-
- Pesce A, De Sanctis D, Nardini M, Dewilde S, Moens L, Hankeln T, Burmester T, Ascenzi P, Bolognesi M. Reversible hexa- to penta-coordination of the heme Fe atom modulates ligand binding properties of neuroglobin and cytoglobin. IUBMB Life. 2004;56:657–64. - PubMed
-
- Genova ML, Lenaz G. Functional role of mitochondrial respiratory supercomplexes. Biochim Biophys Acta. 2014;1837:427–43. - PubMed
-
- Kagan VE, Tyurin VA, Jiang J, Tyurina YY, Ritov VB, Amoscato AA, Osipov AN, Belikova NA, Kapralov AA, Kini V, Vlasova II, Zhao Q, Zou M, Di P, Svistunenko DA, Kurnikov IV, Borisenko GG. Cytochrome c acts as a cardiolipin oxygenase required for release of proapoptotic factors. Nat Chem Biol. 2005;1:223–32. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA165065/CA/NCI NIH HHS/United States
- R01 GM097082/GM/NIGMS NIH HHS/United States
- U19 AI068021/AI/NIAID NIH HHS/United States
- R37 NS061817/NS/NINDS NIH HHS/United States
- R01 NS076511/NS/NINDS NIH HHS/United States
- ES020693/ES/NIEHS NIH HHS/United States
- NS076511/NS/NINDS NIH HHS/United States
- U19AI068021/AI/NIAID NIH HHS/United States
- R01 NS061817/NS/NINDS NIH HHS/United States
- CA165065/CA/NCI NIH HHS/United States
- P01 HL114453/HL/NHLBI NIH HHS/United States
- GM097082/GM/NIGMS NIH HHS/United States
- T32 HL007563/HL/NHLBI NIH HHS/United States
- NS061817/NS/NINDS NIH HHS/United States
- R01 ES020693/ES/NIEHS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

