High-throughput identification of genotype-specific cancer vulnerabilities in mixtures of barcoded tumor cell lines
- PMID: 26928769
- PMCID: PMC5508574
- DOI: 10.1038/nbt.3460
High-throughput identification of genotype-specific cancer vulnerabilities in mixtures of barcoded tumor cell lines
Abstract
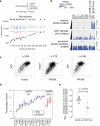
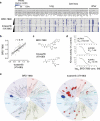
Hundreds of genetically characterized cell lines are available for the discovery of genotype-specific cancer vulnerabilities. However, screening large numbers of compounds against large numbers of cell lines is currently impractical, and such experiments are often difficult to control. Here we report a method called PRISM that allows pooled screening of mixtures of cancer cell lines by labeling each cell line with 24-nucleotide barcodes. PRISM revealed the expected patterns of cell killing seen in conventional (unpooled) assays. In a screen of 102 cell lines across 8,400 compounds, PRISM led to the identification of BRD-7880 as a potent and highly specific inhibitor of aurora kinases B and C. Cell line pools also efficiently formed tumors as xenografts, and PRISM recapitulated the expected pattern of erlotinib sensitivity in vivo.
Figures

References
-
- Sharma SV, Haber DA, Settleman J. Cell line-based platforms to evaluate the therapeutic efficacy of candidate anticancer agents. Nature reviews. Cancer. 2010;10:241–253. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

